| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Indoleamine 2,3-dioxygenase 1 |
|---|
| Ligand | BDBM50030791 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1550111 (CHEMBL3756253) |
|---|
| IC50 | 3000±n/a nM |
|---|
| Citation |  Peng, YH; Ueng, SH; Tseng, CT; Hung, MS; Song, JS; Wu, JS; Liao, FY; Fan, YS; Wu, MH; Hsiao, WC; Hsueh, CC; Lin, SY; Cheng, CY; Tu, CH; Lee, LC; Cheng, MF; Shia, KS; Shih, C; Wu, SY Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1. J Med Chem59:282-93 (2016) [PubMed] Article Peng, YH; Ueng, SH; Tseng, CT; Hung, MS; Song, JS; Wu, JS; Liao, FY; Fan, YS; Wu, MH; Hsiao, WC; Hsueh, CC; Lin, SY; Cheng, CY; Tu, CH; Lee, LC; Cheng, MF; Shia, KS; Shih, C; Wu, SY Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1. J Med Chem59:282-93 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Indoleamine 2,3-dioxygenase 1 |
|---|
| Name: | Indoleamine 2,3-dioxygenase 1 |
|---|
| Synonyms: | I23O1_HUMAN | IDO | IDO-1 | IDO1 | INDO | Indoleamine 2,3-Dioxygenasae (IDO) | Indoleamine 2,3-dioxygenase | Indoleamine-pyrrole 2,3-dioxygenase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 45330.80 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P14902 |
|---|
| Residue: | 403 |
|---|
| Sequence: | MAHAMENSWTISKEYHIDEEVGFALPNPQENLPDFYNDWMFIAKHLPDLIESGQLRERVE
KLNMLSIDHLTDHKSQRLARLVLGCITMAYVWGKGHGDVRKVLPRNIAVPYCQLSKKLEL
PPILVYADCVLANWKKKDPNKPLTYENMDVLFSFRDGDCSKGFFLVSLLVEIAAASAIKV
IPTVFKAMQMQERDTLLKALLEIASCLEKALQVFHQIHDHVNPKAFFSVLRIYLSGWKGN
PQLSDGLVYEGFWEDPKEFAGGSAGQSSVFQCFDVLLGIQQTAGGGHAAQFLQDMRRYMP
PAHRNFLCSLESNPSVREFVLSKGDAGLREAYDACVKALVSLRSYHLQIVTKYILIPASQ
QPKENKTSEDPSKLEAKGTGGTDLMNFLKTVRSTTEKSLLKEG
|
|
|
|---|
| BDBM50030791 |
|---|
| n/a |
|---|
| Name | BDBM50030791 |
|---|
| Synonyms: | CHEMBL3342402 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H16N4O3S2 |
|---|
| Mol. Mass. | 424.496 |
|---|
| SMILES | Cc1ccc(cc1)-c1csc2nnc(SCC(=O)Nc3ccc4OCOc4c3)n12 |
|---|
| Structure | 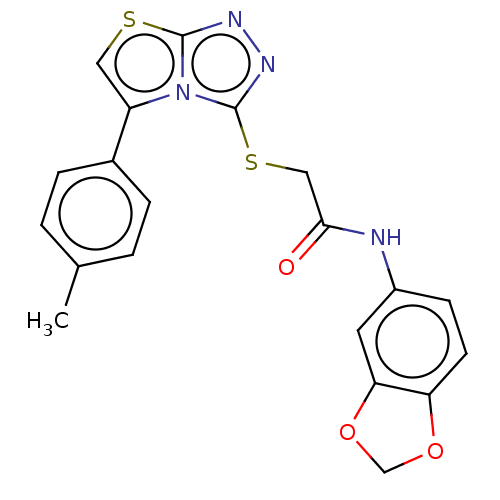
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Peng, YH; Ueng, SH; Tseng, CT; Hung, MS; Song, JS; Wu, JS; Liao, FY; Fan, YS; Wu, MH; Hsiao, WC; Hsueh, CC; Lin, SY; Cheng, CY; Tu, CH; Lee, LC; Cheng, MF; Shia, KS; Shih, C; Wu, SY Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1. J Med Chem59:282-93 (2016) [PubMed] Article
Peng, YH; Ueng, SH; Tseng, CT; Hung, MS; Song, JS; Wu, JS; Liao, FY; Fan, YS; Wu, MH; Hsiao, WC; Hsueh, CC; Lin, SY; Cheng, CY; Tu, CH; Lee, LC; Cheng, MF; Shia, KS; Shih, C; Wu, SY Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1. J Med Chem59:282-93 (2016) [PubMed] Article