| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lysine-specific demethylase 6B |
|---|
| Ligand | BDBM60875 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1548620 (CHEMBL3757620) |
|---|
| IC50 | 150±n/a nM |
|---|
| Citation |  Hu, J; Wang, X; Chen, L; Huang, M; Tang, W; Zuo, J; Liu, YC; Shi, Z; Liu, R; Shen, J; Xiong, B Design and discovery of new pyrimidine coupled nitrogen aromatic rings as chelating groups of JMJD3 inhibitors. Bioorg Med Chem Lett26:721-5 (2016) [PubMed] Article Hu, J; Wang, X; Chen, L; Huang, M; Tang, W; Zuo, J; Liu, YC; Shi, Z; Liu, R; Shen, J; Xiong, B Design and discovery of new pyrimidine coupled nitrogen aromatic rings as chelating groups of JMJD3 inhibitors. Bioorg Med Chem Lett26:721-5 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lysine-specific demethylase 6B |
|---|
| Name: | Lysine-specific demethylase 6B |
|---|
| Synonyms: | JMJD3 | JmjC domain-containing protein 3 | Jumonji domain-containing protein 3 | KDM6B | KDM6B_HUMAN | KIAA0346 | Lysine demethylase 6B | Lysine-specific demethylase 6B | Lysine-specific demethylase 6B (KDM6B(JMJD3)) | Lysine-specific demethylase 6B (KDM6B) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 176672.55 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15054 |
|---|
| Residue: | 1643 |
|---|
| Sequence: | MHRAVDPPGARAAREAFALGGLSCAGAWSSCPPHPPPRSAWLPGGRCSASIGQPPLPAPL
PPSHGSSSGHPSKPYYAPGAPTPRPLHGKLESLHGCVQALLREPAQPGLWEQLGQLYESE
HDSEEATRCYHSALRYGGSFAELGPRIGRLQQAQLWNFHTGSCQHRAKVLPPLEQVWNLL
HLEHKRNYGAKRGGPPVKRAAEPPVVQPVPPAALSGPSGEEGLSPGGKRRRGCNSEQTGL
PPGLPLPPPPLPPPPPPPPPPPPPLPGLATSPPFQLTKPGLWSTLHGDAWGPERKGSAPP
ERQEQRHSLPHPYPYPAPAYTAHPPGHRLVPAAPPGPGPRPPGAESHGCLPATRPPGSDL
RESRVQRSRMDSSVSPAATTACVPYAPSRPPGLPGTTTSSSSSSSSNTGLRGVEPNPGIP
GADHYQTPALEVSHHGRLGPSAHSSRKPFLGAPAATPHLSLPPGPSSPPPPPCPRLLRPP
PPPAWLKGPACRAAREDGEILEELFFGTEGPPRPAPPPLPHREGFLGPPASRFSVGTQDS
HTPPTPPTPTTSSSNSNSGSHSSSPAGPVSFPPPPYLARSIDPLPRPPSPAQNPQDPPLV
PLTLALPPAPPSSCHQNTSGSFRRPESPRPRVSFPKTPEVGPGPPPGPLSKAPQPVPPGV
GELPARGPRLFDFPPTPLEDQFEEPAEFKILPDGLANIMKMLDESIRKEEEQQQHEAGVA
PQPPLKEPFASLQSPFPTDTAPTTTAPAVAVTTTTTTTTTTTATQEEEKKPPPALPPPPP
LAKFPPPSQPQPPPPPPPSPASLLKSLASVLEGQKYCYRGTGAAVSTRPGPLPTTQYSPG
PPSGATALPPTSAAPSAQGSPQPSASSSSQFSTSGGPWARERRAGEEPVPGPMTPTQPPP
PLSLPPARSESEVLEEISRACETLVERVGRSATDPADPVDTAEPADSGTERLLPPAQAKE
EAGGVAAVSGSCKRRQKEHQKEHRRHRRACKDSVGRRPREGRAKAKAKVPKEKSRRVLGN
LDLQSEEIQGREKSRPDLGGASKAKPPTAPAPPSAPAPSAQPTPPSASVPGKKAREEAPG
PPGVSRADMLKLRSLSEGPPKELKIRLIKVESGDKETFIASEVEERRLRMADLTISHCAA
DVVRASRNAKVKGKFRESYLSPAQSVKPKINTEEKLPREKLNPPTPSIYLESKRDAFSPV
LLQFCTDPRNPITVIRGLAGSLRLNLGLFSTKTLVEASGEHTVEVRTQVQQPSDENWDLT
GTRQIWPCESSRSHTTIAKYAQYQASSFQESLQEEKESEDEESEEPDSTTGTPPSSAPDP
KNHHIIKFGTNIDLSDAKRWKPQLQELLKLPAFMRVTSTGNMLSHVGHTILGMNTVQLYM
KVPGSRTPGHQENNNFCSVNINIGPGDCEWFAVHEHYWETISAFCDRHGVDYLTGSWWPI
LDDLYASNIPVYRFVQRPGDLVWINAGTVHWVQATGWCNNIAWNVGPLTAYQYQLALERY
EWNEVKNVKSIVPMIHVSWNVARTVKISDPDLFKMIKFCLLQSMKHCQVQRESLVRAGKK
IAYQGRVKDEPAYYCNECDVEVFNILFVTSENGSRNTYLVHCEGCARRRSAGLQGVVVLE
QYRTEELAQAYDAFTLAPASTSR
|
|
|
|---|
| BDBM60875 |
|---|
| n/a |
|---|
| Name | BDBM60875 |
|---|
| Synonyms: | 3-((6-(4,5-Dihydro-1H-benzo[d]azepin-3(2H)-yl)-2-(pyridin-2-yl)pyrimidin-4-yl)amino)propanoic acid | 3-{[2-(pyridin-2-yl)-6-(2,3,4,5-tetrahydro-1H-3-benzazepin-3-yl)pyrimidin-4-yl]amino}propanoic acid | GSK J1 | GSK-J1 | GSKJ1 |
|---|
| Type | n/a |
|---|
| Emp. Form. | C22H23N5O2 |
|---|
| Mol. Mass. | 389.4503 |
|---|
| SMILES | OC(=O)CCNc1cc(nc(n1)-c1ccccn1)N1CCc2ccccc2CC1 |
|---|
| Structure | 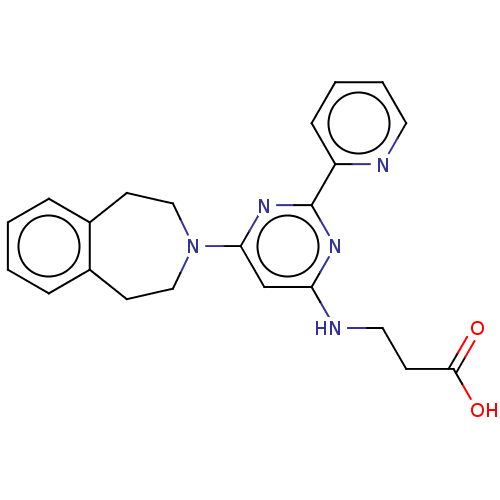
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hu, J; Wang, X; Chen, L; Huang, M; Tang, W; Zuo, J; Liu, YC; Shi, Z; Liu, R; Shen, J; Xiong, B Design and discovery of new pyrimidine coupled nitrogen aromatic rings as chelating groups of JMJD3 inhibitors. Bioorg Med Chem Lett26:721-5 (2016) [PubMed] Article
Hu, J; Wang, X; Chen, L; Huang, M; Tang, W; Zuo, J; Liu, YC; Shi, Z; Liu, R; Shen, J; Xiong, B Design and discovery of new pyrimidine coupled nitrogen aromatic rings as chelating groups of JMJD3 inhibitors. Bioorg Med Chem Lett26:721-5 (2016) [PubMed] Article