| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Diacylglycerol lipase-alpha |
|---|
| Ligand | BDBM50048620 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1553499 (CHEMBL3768670) |
|---|
| IC50 | 18±n/a nM |
|---|
| Citation |  Chupak, LS; Zheng, X; Hu, S; Huang, Y; Ding, M; Lewis, MA; Westphal, RS; Blat, Y; McClure, A; Gentles, RG Structure activity relationship studies on chemically non-reactive glycine sulfonamide inhibitors of diacylglycerol lipase. Bioorg Med Chem24:1455-68 (2016) [PubMed] Article Chupak, LS; Zheng, X; Hu, S; Huang, Y; Ding, M; Lewis, MA; Westphal, RS; Blat, Y; McClure, A; Gentles, RG Structure activity relationship studies on chemically non-reactive glycine sulfonamide inhibitors of diacylglycerol lipase. Bioorg Med Chem24:1455-68 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Diacylglycerol lipase-alpha |
|---|
| Name: | Diacylglycerol lipase-alpha |
|---|
| Synonyms: | C11orf11 | DAGLA | DGLA_HUMAN | KIAA0659 | NSDDR | Sn1-specific diacylglycerol lipase alpha |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 114947.74 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1449471 |
|---|
| Residue: | 1042 |
|---|
| Sequence: | MPGIVVFRRRWSVGSDDLVLPAIFLFLLHTTWFVILSVVLFGLVYNPHEACSLNLVDHGR
GYLGILLSCMIAEMAIIWLSMRGGILYTEPRDSMQYVLYVRLAILVIEFIYAIVGIVWLT
QYYTSCNDLTAKNVTLGMVVCNWVVILSVCITVLCVFDPTGRTFVKLRATKRRQRNLRTY
NLRHRLEEGQATSWSRRLKVFLCCTRTKDSQSDAYSEIAYLFAEFFRDLDIVPSDIIAGL
VLLRQRQRAKRNAVLDEANNDILAFLSGMPVTRNTKYLDLKNSQEMLRYKEVCYYMLFAL
AAYGWPMYLMRKPACGLCQLARSCSCCLCPARPRFAPGVTIEEDNCCGCNAIAIRRHFLD
ENMTAVDIVYTSCHDAVYETPFYVAVDHDKKKVVISIRGTLSPKDALTDLTGDAERLPVE
GHHGTWLGHKGMVLSAEYIKKKLEQEMVLSQAFGRDLGRGTKHYGLIVVGHSLGAGTAAI
LSFLLRPQYPTLKCFAYSPPGGLLSEDAMEYSKEFVTAVVLGKDLVPRIGLSQLEGFRRQ
LLDVLQRSTKPKWRIIVGATKCIPKSELPEEVEVTTLASTRLWTHPSDLTIALSASTPLY
PPGRIIHVVHNHPAEQCCCCEQEEPTYFAIWGDNKAFNEVIISPAMLHEHLPYVVMEGLN
KVLENYNKGKTALLSAAKVMVSPTEVDLTPELIFQQQPLPTGPPMPTGLALELPTADHRN
SSVRSKSQSEMSLEGFSEGRLLSPVVAAAARQDPVELLLLSTQERLAAELQARRAPLATM
ESLSDTESLYSFDSRRSSGFRSIRGSPSLHAVLERDEGHLFYIDPAIPEENPSLSSRTEL
LAADSLSKHSQDTQPLEAALGSGGVTPERPPSAAANDEEEEVGGGGGGPASRGELALHNG
RLGDSPSPQVLEFAEFIDSLFNLDSKSSSFQDLYCMVVPESPTSDYAEGPKSPSQQEILL
RAQFEPNLVPKPPRLFAGSADPSSGISLSPSFPLSSSGELMDLTPTGLSSQECLAADKIR
TSTPTGHGASPAKQDELVISAR
|
|
|
|---|
| BDBM50048620 |
|---|
| n/a |
|---|
| Name | BDBM50048620 |
|---|
| Synonyms: | CHEMBL3319620 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H27NO6S |
|---|
| Mol. Mass. | 481.561 |
|---|
| SMILES | CC1(C)CCc2cc(ccc2O1)S(=O)(=O)N(CC(O)=O)Cc1ccc(Oc2ccccc2)cc1 |
|---|
| Structure | 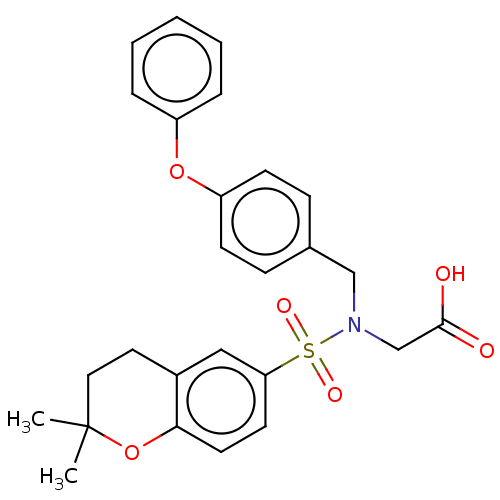
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chupak, LS; Zheng, X; Hu, S; Huang, Y; Ding, M; Lewis, MA; Westphal, RS; Blat, Y; McClure, A; Gentles, RG Structure activity relationship studies on chemically non-reactive glycine sulfonamide inhibitors of diacylglycerol lipase. Bioorg Med Chem24:1455-68 (2016) [PubMed] Article
Chupak, LS; Zheng, X; Hu, S; Huang, Y; Ding, M; Lewis, MA; Westphal, RS; Blat, Y; McClure, A; Gentles, RG Structure activity relationship studies on chemically non-reactive glycine sulfonamide inhibitors of diacylglycerol lipase. Bioorg Med Chem24:1455-68 (2016) [PubMed] Article