| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Polyamine deacetylase HDAC10 |
|---|
| Ligand | BDBM50439674 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1560793 (CHEMBL3778490) |
|---|
| IC50 | 194±n/a nM |
|---|
| Citation |  Yang, Z; Wang, T; Wang, F; Niu, T; Liu, Z; Chen, X; Long, C; Tang, M; Cao, D; Wang, X; Xiang, W; Yi, Y; Ma, L; You, J; Chen, L Discovery of Selective Histone Deacetylase 6 Inhibitors Using the Quinazoline as the Cap for the Treatment of Cancer. J Med Chem59:1455-70 (2016) [PubMed] Article Yang, Z; Wang, T; Wang, F; Niu, T; Liu, Z; Chen, X; Long, C; Tang, M; Cao, D; Wang, X; Xiang, W; Yi, Y; Ma, L; You, J; Chen, L Discovery of Selective Histone Deacetylase 6 Inhibitors Using the Quinazoline as the Cap for the Treatment of Cancer. J Med Chem59:1455-70 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Polyamine deacetylase HDAC10 |
|---|
| Name: | Polyamine deacetylase HDAC10 |
|---|
| Synonyms: | HD10 | HDA10_HUMAN | HDAC10 | Histone deacetylase | Histone deacetylase 10 | Human HDAC10 |
|---|
| Type: | Chromatin regulator; hydrolase; repressor |
|---|
| Mol. Mass.: | 71431.89 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q969S8 |
|---|
| Residue: | 669 |
|---|
| Sequence: | MGTALVYHEDMTATRLLWDDPECEIERPERLTAALDRLRQRGLEQRCLRLSAREASEEEL
GLVHSPEYVSLVRETQVLGKEELQALSGQFDAIYFHPSTFHCARLAAGAGLQLVDAVLTG
AVQNGLALVRPPGHHGQRAAANGFCVFNNVAIAAAHAKQKHGLHRILVVDWDVHHGQGIQ
YLFEDDPSVLYFSWHRYEHGRFWPFLRESDADAVGRGQGLGFTVNLPWNQVGMGNADYVA
AFLHLLLPLAFEFDPELVLVSAGFDSAIGDPEGQMQATPECFAHLTQLLQVLAGGRVCAV
LEGGYHLESLAESVCMTVQTLLGDPAPPLSGPMAPCQSALESIQSARAAQAPHWKSLQQQ
DVTAVPMSPSSHSPEGRPPPLLPGGPVCKAAASAPSSLLDQPCLCPAPSVRTAVALTTPD
ITLVLPPDVIQQEASALREETEAWARPHESLAREEALTALGKLLYLLDGMLDGQVNSGIA
ATPASAAAATLDVAVRRGLSHGAQRLLCVALGQLDRPPDLAHDGRSLWLNIRGKEAAALS
MFHVSTPLPVMTGGFLSCILGLVLPLAYGFQPDLVLVALGPGHGLQGPHAALLAAMLRGL
AGGRVLALLEENSTPQLAGILARVLNGEAPPSLGPSSVASPEDVQALMYLRGQLEPQWKM
LQCHPHLVA
|
|
|
|---|
| BDBM50439674 |
|---|
| n/a |
|---|
| Name | BDBM50439674 |
|---|
| Synonyms: | RICOLINOSTAT | US10858323, Compound 2 | US11207431, Example E | US20230414581, Compound 43 | US8609678, 2-(diphenylamino)-N-(7-(hydroxyamino)-7-oxoheptyl)pyrimidine-5-carboxamide [26] |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H27N5O3 |
|---|
| Mol. Mass. | 433.5029 |
|---|
| SMILES | ONC(=O)CCCCCCNC(=O)c1cnc(nc1)N(c1ccccc1)c1ccccc1 |
|---|
| Structure | 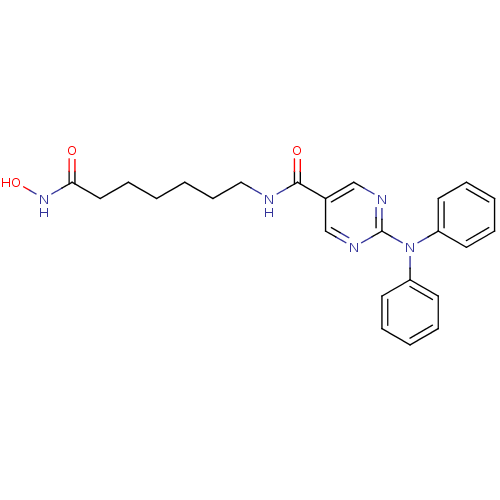
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Yang, Z; Wang, T; Wang, F; Niu, T; Liu, Z; Chen, X; Long, C; Tang, M; Cao, D; Wang, X; Xiang, W; Yi, Y; Ma, L; You, J; Chen, L Discovery of Selective Histone Deacetylase 6 Inhibitors Using the Quinazoline as the Cap for the Treatment of Cancer. J Med Chem59:1455-70 (2016) [PubMed] Article
Yang, Z; Wang, T; Wang, F; Niu, T; Liu, Z; Chen, X; Long, C; Tang, M; Cao, D; Wang, X; Xiang, W; Yi, Y; Ma, L; You, J; Chen, L Discovery of Selective Histone Deacetylase 6 Inhibitors Using the Quinazoline as the Cap for the Treatment of Cancer. J Med Chem59:1455-70 (2016) [PubMed] Article