| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lysine-specific demethylase 6B |
|---|
| Ligand | BDBM50153348 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1562427 (CHEMBL3778163) |
|---|
| IC50 | 2512±n/a nM |
|---|
| Citation |  Westaway, SM; Preston, AG; Barker, MD; Brown, F; Brown, JA; Campbell, M; Chung, CW; Diallo, H; Douault, C; Drewes, G; Eagle, R; Gordon, L; Haslam, C; Hayhow, TG; Humphreys, PG; Joberty, G; Katso, R; Kruidenier, L; Leveridge, M; Liddle, J; Mosley, J; Muelbaier, M; Randle, R; Rioja, I; Rueger, A; Seal, GA; Sheppard, RJ; Singh, O; Taylor, J; Thomas, P; Thomson, D; Wilson, DM; Lee, K; Prinjha, RK Cell Penetrant Inhibitors of the KDM4 and KDM5 Families of Histone Lysine Demethylases. 1. 3-Amino-4-pyridine Carboxylate Derivatives. J Med Chem59:1357-69 (2016) [PubMed] Article Westaway, SM; Preston, AG; Barker, MD; Brown, F; Brown, JA; Campbell, M; Chung, CW; Diallo, H; Douault, C; Drewes, G; Eagle, R; Gordon, L; Haslam, C; Hayhow, TG; Humphreys, PG; Joberty, G; Katso, R; Kruidenier, L; Leveridge, M; Liddle, J; Mosley, J; Muelbaier, M; Randle, R; Rioja, I; Rueger, A; Seal, GA; Sheppard, RJ; Singh, O; Taylor, J; Thomas, P; Thomson, D; Wilson, DM; Lee, K; Prinjha, RK Cell Penetrant Inhibitors of the KDM4 and KDM5 Families of Histone Lysine Demethylases. 1. 3-Amino-4-pyridine Carboxylate Derivatives. J Med Chem59:1357-69 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lysine-specific demethylase 6B |
|---|
| Name: | Lysine-specific demethylase 6B |
|---|
| Synonyms: | JMJD3 | JmjC domain-containing protein 3 | Jumonji domain-containing protein 3 | KDM6B | KDM6B_HUMAN | KIAA0346 | Lysine demethylase 6B | Lysine-specific demethylase 6B | Lysine-specific demethylase 6B (KDM6B(JMJD3)) | Lysine-specific demethylase 6B (KDM6B) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 176672.55 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15054 |
|---|
| Residue: | 1643 |
|---|
| Sequence: | MHRAVDPPGARAAREAFALGGLSCAGAWSSCPPHPPPRSAWLPGGRCSASIGQPPLPAPL
PPSHGSSSGHPSKPYYAPGAPTPRPLHGKLESLHGCVQALLREPAQPGLWEQLGQLYESE
HDSEEATRCYHSALRYGGSFAELGPRIGRLQQAQLWNFHTGSCQHRAKVLPPLEQVWNLL
HLEHKRNYGAKRGGPPVKRAAEPPVVQPVPPAALSGPSGEEGLSPGGKRRRGCNSEQTGL
PPGLPLPPPPLPPPPPPPPPPPPPLPGLATSPPFQLTKPGLWSTLHGDAWGPERKGSAPP
ERQEQRHSLPHPYPYPAPAYTAHPPGHRLVPAAPPGPGPRPPGAESHGCLPATRPPGSDL
RESRVQRSRMDSSVSPAATTACVPYAPSRPPGLPGTTTSSSSSSSSNTGLRGVEPNPGIP
GADHYQTPALEVSHHGRLGPSAHSSRKPFLGAPAATPHLSLPPGPSSPPPPPCPRLLRPP
PPPAWLKGPACRAAREDGEILEELFFGTEGPPRPAPPPLPHREGFLGPPASRFSVGTQDS
HTPPTPPTPTTSSSNSNSGSHSSSPAGPVSFPPPPYLARSIDPLPRPPSPAQNPQDPPLV
PLTLALPPAPPSSCHQNTSGSFRRPESPRPRVSFPKTPEVGPGPPPGPLSKAPQPVPPGV
GELPARGPRLFDFPPTPLEDQFEEPAEFKILPDGLANIMKMLDESIRKEEEQQQHEAGVA
PQPPLKEPFASLQSPFPTDTAPTTTAPAVAVTTTTTTTTTTTATQEEEKKPPPALPPPPP
LAKFPPPSQPQPPPPPPPSPASLLKSLASVLEGQKYCYRGTGAAVSTRPGPLPTTQYSPG
PPSGATALPPTSAAPSAQGSPQPSASSSSQFSTSGGPWARERRAGEEPVPGPMTPTQPPP
PLSLPPARSESEVLEEISRACETLVERVGRSATDPADPVDTAEPADSGTERLLPPAQAKE
EAGGVAAVSGSCKRRQKEHQKEHRRHRRACKDSVGRRPREGRAKAKAKVPKEKSRRVLGN
LDLQSEEIQGREKSRPDLGGASKAKPPTAPAPPSAPAPSAQPTPPSASVPGKKAREEAPG
PPGVSRADMLKLRSLSEGPPKELKIRLIKVESGDKETFIASEVEERRLRMADLTISHCAA
DVVRASRNAKVKGKFRESYLSPAQSVKPKINTEEKLPREKLNPPTPSIYLESKRDAFSPV
LLQFCTDPRNPITVIRGLAGSLRLNLGLFSTKTLVEASGEHTVEVRTQVQQPSDENWDLT
GTRQIWPCESSRSHTTIAKYAQYQASSFQESLQEEKESEDEESEEPDSTTGTPPSSAPDP
KNHHIIKFGTNIDLSDAKRWKPQLQELLKLPAFMRVTSTGNMLSHVGHTILGMNTVQLYM
KVPGSRTPGHQENNNFCSVNINIGPGDCEWFAVHEHYWETISAFCDRHGVDYLTGSWWPI
LDDLYASNIPVYRFVQRPGDLVWINAGTVHWVQATGWCNNIAWNVGPLTAYQYQLALERY
EWNEVKNVKSIVPMIHVSWNVARTVKISDPDLFKMIKFCLLQSMKHCQVQRESLVRAGKK
IAYQGRVKDEPAYYCNECDVEVFNILFVTSENGSRNTYLVHCEGCARRRSAGLQGVVVLE
QYRTEELAQAYDAFTLAPASTSR
|
|
|
|---|
| BDBM50153348 |
|---|
| n/a |
|---|
| Name | BDBM50153348 |
|---|
| Synonyms: | CHEMBL3774403 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H14N2O2 |
|---|
| Mol. Mass. | 194.2304 |
|---|
| SMILES | CCCCNc1cnccc1C(O)=O |
|---|
| Structure | 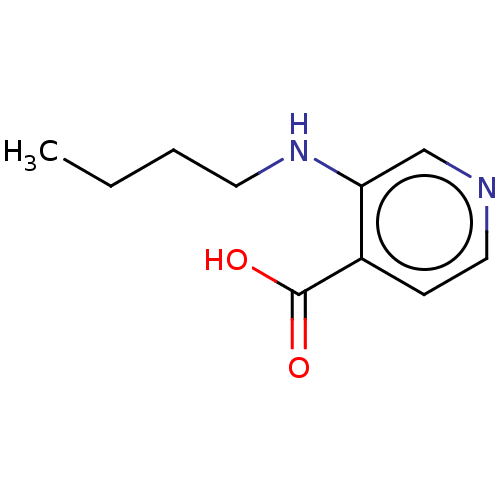
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Westaway, SM; Preston, AG; Barker, MD; Brown, F; Brown, JA; Campbell, M; Chung, CW; Diallo, H; Douault, C; Drewes, G; Eagle, R; Gordon, L; Haslam, C; Hayhow, TG; Humphreys, PG; Joberty, G; Katso, R; Kruidenier, L; Leveridge, M; Liddle, J; Mosley, J; Muelbaier, M; Randle, R; Rioja, I; Rueger, A; Seal, GA; Sheppard, RJ; Singh, O; Taylor, J; Thomas, P; Thomson, D; Wilson, DM; Lee, K; Prinjha, RK Cell Penetrant Inhibitors of the KDM4 and KDM5 Families of Histone Lysine Demethylases. 1. 3-Amino-4-pyridine Carboxylate Derivatives. J Med Chem59:1357-69 (2016) [PubMed] Article
Westaway, SM; Preston, AG; Barker, MD; Brown, F; Brown, JA; Campbell, M; Chung, CW; Diallo, H; Douault, C; Drewes, G; Eagle, R; Gordon, L; Haslam, C; Hayhow, TG; Humphreys, PG; Joberty, G; Katso, R; Kruidenier, L; Leveridge, M; Liddle, J; Mosley, J; Muelbaier, M; Randle, R; Rioja, I; Rueger, A; Seal, GA; Sheppard, RJ; Singh, O; Taylor, J; Thomas, P; Thomson, D; Wilson, DM; Lee, K; Prinjha, RK Cell Penetrant Inhibitors of the KDM4 and KDM5 Families of Histone Lysine Demethylases. 1. 3-Amino-4-pyridine Carboxylate Derivatives. J Med Chem59:1357-69 (2016) [PubMed] Article