| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone lysine demethylase PHF8 |
|---|
| Ligand | BDBM50155505 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_1565735 |
|---|
| IC50 | <250±n/a nM |
|---|
| Citation |  Abdel-Magid, AF Inhibition of Histone Demethylases Offers a Novel and Promising Approach for the Treatment of Cancer and Other Diseases. ACS Med Chem Lett7:128-30 (2016) [PubMed] Article Abdel-Magid, AF Inhibition of Histone Demethylases Offers a Novel and Promising Approach for the Treatment of Cancer and Other Diseases. ACS Med Chem Lett7:128-30 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone lysine demethylase PHF8 |
|---|
| Name: | Histone lysine demethylase PHF8 |
|---|
| Synonyms: | Histone lysine demethylase PHF8 | Histone lysine demethylase PHF8 (PHF8) | KIAA1111 | Lysine-specific demethylase 7A/7B | PHD finger protein 8 | PHF8 | PHF8_HUMAN | ZNF422 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 117889.91 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9UPP1 |
|---|
| Residue: | 1060 |
|---|
| Sequence: | MNRSRAIVQRGRVLPPPAPLDTTNLAGRRTLQGRAKMASVPVYCLCRLPYDVTRFMIECD
MCQDWFHGSCVGVEEEKAADIDLYHCPNCEVLHGPSIMKKRRGSSKGHDTHKGKPVKTGS
PTFVRELRSRTFDSSDEVILKPTGNQLTVEFLEENSFSVPILVLKKDGLGMTLPSPSFTV
RDVEHYVGSDKEIDVIDVTRQADCKMKLGDFVKYYYSGKREKVLNVISLEFSDTRLSNLV
ETPKIVRKLSWVENLWPEECVFERPNVQKYCLMSVRDSYTDFHIDFGGTSVWYHVLKGEK
IFYLIRPTNANLTLFECWSSSSNQNEMFFGDQVDKCYKCSVKQGQTLFIPTGWIHAVLTP
VDCLAFGGNFLHSLNIEMQLKAYEIEKRLSTADLFRFPNFETICWYVGKHILDIFRGLRE
NRRHPASYLVHGGKALNLAFRAWTRKEALPDHEDEIPETVRTVQLIKDLAREIRLVEDIF
QQNVGKTSNIFGLQRIFPAGSIPLTRPAHSTSVSMSRLSLPSKNGSKKKGLKPKELFKKA
ERKGKESSALGPAGQLSYNLMDTYSHQALKTGSFQKAKFNITGACLNDSDDDSPDLDLDG
NESPLALLMSNGSTKRVKSLSKSRRTKIAKKVDKARLMAEQVMEDEFDLDSDDELQIDER
LGKEKATLIIRPKFPRKLPRAKPCSDPNRVREPGEVEFDIEEDYTTDEDMVEGVEGKLGN
GSGAGGILDLLKASRQVGGPDYAALTEAPASPSTQEAIQGMLCMANLQSSSSSPATSSLQ
AWWTGGQDRSSGSSSSGLGTVSNSPASQRTPGKRPIKRPAYWRTESEEEEENASLDEQDS
LGACFKDAEYIYPSLESDDDDPALKSRPKKKKNSDDAPWSPKARVTPTLPKQDRPVREGT
RVASIETGLAAAAAKLAQQELQKAQKKKYIKKKPLLKEVEQPRPQDSNLSLTVPAPTVAA
TPQLVTSSSPLPPPEPKQEALSGSLADHEYTARPNAFGMAQANRSTTPMAPGVFLTQRRP
SVGSQSNQAGQGKRPKKGLATAKQRLGRILKIHRNGKLLL
|
|
|
|---|
| BDBM50155505 |
|---|
| n/a |
|---|
| Name | BDBM50155505 |
|---|
| Synonyms: | CHEMBL3781414 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H19N3O3 |
|---|
| Mol. Mass. | 277.319 |
|---|
| SMILES | CCN1CCC(CNCc2cc(ccn2)C(O)=O)C1=O |
|---|
| Structure | 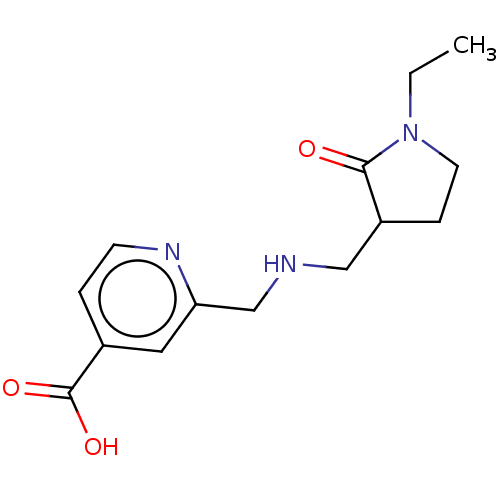
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Abdel-Magid, AF Inhibition of Histone Demethylases Offers a Novel and Promising Approach for the Treatment of Cancer and Other Diseases. ACS Med Chem Lett7:128-30 (2016) [PubMed] Article
Abdel-Magid, AF Inhibition of Histone Demethylases Offers a Novel and Promising Approach for the Treatment of Cancer and Other Diseases. ACS Med Chem Lett7:128-30 (2016) [PubMed] Article