| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 |
|---|
| Ligand | BDBM60622 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1566102 (CHEMBL3791986) |
|---|
| IC50 | >10000±n/a nM |
|---|
| Citation |  Brindisi, M; Maramai, S; Gemma, S; Brogi, S; Grillo, A; Di Cesare Mannelli, L; Gabellieri, E; Lamponi, S; Saponara, S; Gorelli, B; Tedesco, D; Bonfiglio, T; Landry, C; Jung, KM; Armirotti, A; Luongo, L; Ligresti, A; Piscitelli, F; Bertucci, C; Dehouck, MP; Campiani, G; Maione, S; Ghelardini, C; Pittaluga, A; Piomelli, D; Di Marzo, V; Butini, S Development and Pharmacological Characterization of Selective Blockers of 2-Arachidonoyl Glycerol Degradation with Efficacy in Rodent Models of Multiple Sclerosis and Pain. J Med Chem59:2612-32 (2016) [PubMed] Article Brindisi, M; Maramai, S; Gemma, S; Brogi, S; Grillo, A; Di Cesare Mannelli, L; Gabellieri, E; Lamponi, S; Saponara, S; Gorelli, B; Tedesco, D; Bonfiglio, T; Landry, C; Jung, KM; Armirotti, A; Luongo, L; Ligresti, A; Piscitelli, F; Bertucci, C; Dehouck, MP; Campiani, G; Maione, S; Ghelardini, C; Pittaluga, A; Piomelli, D; Di Marzo, V; Butini, S Development and Pharmacological Characterization of Selective Blockers of 2-Arachidonoyl Glycerol Degradation with Efficacy in Rodent Models of Multiple Sclerosis and Pain. J Med Chem59:2612-32 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 |
|---|
| Name: | Fatty-acid amide hydrolase 1 |
|---|
| Synonyms: | Anandamide amidohydrolase | Anandamide amidohydrolase 1 | FAAH1_MOUSE | Faah | Faah1 | Fatty Acid Amide Hydrolase | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Oleamide hydrolase 1 |
|---|
| Type: | Hydrolase; single-pass membrane protein; homodimer |
|---|
| Mol. Mass.: | 63227.28 |
|---|
| Organism: | Mus musculus (mouse) |
|---|
| Description: | Mouse brain membranes were used in the assay. |
|---|
| Residue: | 579 |
|---|
| Sequence: | MVLSEVWTALSGLSGVCLACSLLSAAVVLRWTRSQTARGAVTRARQKQRAGLETMDKAVQ
RFRLQNPDLDSEALLALPLLQLVQKLQSGELSPEAVLFTYLGKAWEVNKGTNCVTSYLTD
CETQLSQAPRQGLLYGVPVSLKECFSYKGHASTLGLSLNEGVTSESDCVVVQVLKLQGAV
PFVHTNVPQSMLSYDCSNPLFGQTMNPWKPSKSPGGSSGGEGALIGSGGSPLGLGTDIGG
SIRFPSAFCGICGLKPTGNRLSKSGLKSCVYGQTAVQLSVGPMARDVDSLALCMKALLCE
DLFRLDSTIPPLPFREEIYRSSRPLRVGYYETDNYTMPTPAMRRAVMETKQSLEAAGHTL
VPFLPNNIPYALEVLSAGGLFSDGGCSFLQNFKGDFVDPCLGDLVLVLKLPRWFKKLLSF
LLKPLFPRLAAFLNSMCPRSAEKLWELQHEIEMYRQSVIAQWKAMNLDVVLTPMLGPALD
LNTPGRATGAISYTVLYNCLDFPAGVVPVTTVTAEDDAQMEHYKGYFGDMWDNILKKGMK
KGIGLPVAVQCVALPWQEELCLRFMREVERLMTPEKRPS
|
|
|
|---|
| BDBM60622 |
|---|
| n/a |
|---|
| Name | BDBM60622 |
|---|
| Synonyms: | BDBM50300355 | US11753371, Compound JZL-184 | US9133148, A |
|---|
| Type | n/a |
|---|
| Emp. Form. | C27H24N2O9 |
|---|
| Mol. Mass. | 520.4875 |
|---|
| SMILES | OC(C1CCN(CC1)C(=O)Oc1ccc(cc1)[N+]([O-])=O)(c1ccc2OCOc2c1)c1ccc2OCOc2c1 |
|---|
| Structure | 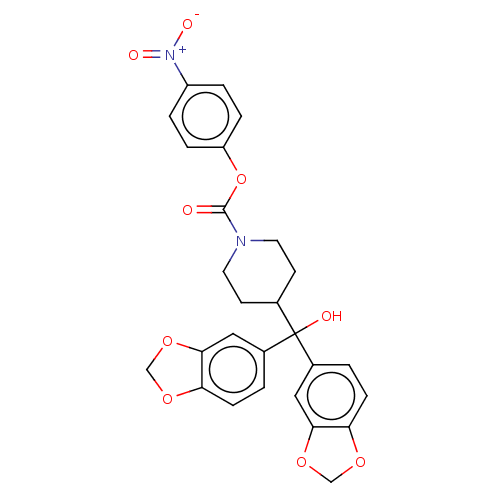
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Brindisi, M; Maramai, S; Gemma, S; Brogi, S; Grillo, A; Di Cesare Mannelli, L; Gabellieri, E; Lamponi, S; Saponara, S; Gorelli, B; Tedesco, D; Bonfiglio, T; Landry, C; Jung, KM; Armirotti, A; Luongo, L; Ligresti, A; Piscitelli, F; Bertucci, C; Dehouck, MP; Campiani, G; Maione, S; Ghelardini, C; Pittaluga, A; Piomelli, D; Di Marzo, V; Butini, S Development and Pharmacological Characterization of Selective Blockers of 2-Arachidonoyl Glycerol Degradation with Efficacy in Rodent Models of Multiple Sclerosis and Pain. J Med Chem59:2612-32 (2016) [PubMed] Article
Brindisi, M; Maramai, S; Gemma, S; Brogi, S; Grillo, A; Di Cesare Mannelli, L; Gabellieri, E; Lamponi, S; Saponara, S; Gorelli, B; Tedesco, D; Bonfiglio, T; Landry, C; Jung, KM; Armirotti, A; Luongo, L; Ligresti, A; Piscitelli, F; Bertucci, C; Dehouck, MP; Campiani, G; Maione, S; Ghelardini, C; Pittaluga, A; Piomelli, D; Di Marzo, V; Butini, S Development and Pharmacological Characterization of Selective Blockers of 2-Arachidonoyl Glycerol Degradation with Efficacy in Rodent Models of Multiple Sclerosis and Pain. J Med Chem59:2612-32 (2016) [PubMed] Article