| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 7 |
|---|
| Ligand | BDBM50188637 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1590568 (CHEMBL3829037) |
|---|
| IC50 | >1000±n/a nM |
|---|
| Citation |  Chen, Y; Wang, X; Xiang, W; He, L; Tang, M; Wang, F; Wang, T; Yang, Z; Yi, Y; Wang, H; Niu, T; Zheng, L; Lei, L; Li, X; Song, H; Chen, L Development of Purine-Based Hydroxamic Acid Derivatives: Potent Histone Deacetylase Inhibitors with Marked in Vitro and in Vivo Antitumor Activities. J Med Chem59:5488-504 (2016) [PubMed] Article Chen, Y; Wang, X; Xiang, W; He, L; Tang, M; Wang, F; Wang, T; Yang, Z; Yi, Y; Wang, H; Niu, T; Zheng, L; Lei, L; Li, X; Song, H; Chen, L Development of Purine-Based Hydroxamic Acid Derivatives: Potent Histone Deacetylase Inhibitors with Marked in Vitro and in Vivo Antitumor Activities. J Med Chem59:5488-504 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 7 |
|---|
| Name: | Histone deacetylase 7 |
|---|
| Synonyms: | HD7 | HDAC7 | HDAC7A | HDAC7_HUMAN | Histone acetylase 7 (HDAC7) | Histone deacetylase 7A | Human HDAC7 |
|---|
| Type: | Chromatin regulator; hydrolase; repressor |
|---|
| Mol. Mass.: | 102942.62 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q8WUI4 |
|---|
| Residue: | 952 |
|---|
| Sequence: | MDLRVGQRPPVEPPPEPTLLALQRPQRLHHHLFLAGLQQQRSVEPMRLSMDTPMPELQVG
PQEQELRQLLHKDKSKRSAVASSVVKQKLAEVILKKQQAALERTVHPNSPGIPYRTLEPL
ETEGATRSMLSSFLPPVPSLPSDPPEHFPLRKTVSEPNLKLRYKPKKSLERRKNPLLRKE
SAPPSLRRRPAETLGDSSPSSSSTPASGCSSPNDSEHGPNPILGSEALLGQRLRLQETSV
APFALPTVSLLPAITLGLPAPARADSDRRTHPTLGPRGPILGSPHTPLFLPHGLEPEAGG
TLPSRLQPILLLDPSGSHAPLLTVPGLGPLPFHFAQSLMTTERLSGSGLHWPLSRTRSEP
LPPSATAPPPPGPMQPRLEQLKTHVQVIKRSAKPSEKPRLRQIPSAEDLETDGGGPGQVV
DDGLEHRELGHGQPEARGPAPLQQHPQVLLWEQQRLAGRLPRGSTGDTVLLPLAQGGHRP
LSRAQSSPAAPASLSAPEPASQARVLSSSETPARTLPFTTGLIYDSVMLKHQCSCGDNSR
HPEHAGRIQSIWSRLQERGLRSQCECLRGRKASLEELQSVHSERHVLLYGTNPLSRLKLD
NGKLAGLLAQRMFVMLPCGGVGVDTDTIWNELHSSNAARWAAGSVTDLAFKVASRELKNG
FAVVRPPGHHADHSTAMGFCFFNSVAIACRQLQQQSKASKILIVDWDVHHGNGTQQTFYQ
DPSVLYISLHRHDDGNFFPGSGAVDEVGAGSGEGFNVNVAWAGGLDPPMGDPEYLAAFRI
VVMPIAREFSPDLVLVSAGFDAAEGHPAPLGGYHVSAKCFGYMTQQLMNLAGGAVVLALE
GGHDLTAICDASEACVAALLGNRVDPLSEEGWKQKPNLNAIRSLEAVIRVHSKYWGCMQR
LASCPDSWVPRVPGADKEEVEAVTALASLSVGILAEDRPSEQLVEEEEPMNL
|
|
|
|---|
| BDBM50188637 |
|---|
| n/a |
|---|
| Name | BDBM50188637 |
|---|
| Synonyms: | CHEMBL3827814 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H26N10O3 |
|---|
| Mol. Mass. | 490.5177 |
|---|
| SMILES | CN(Cc1nc2c(nc(nc2n1C)-c1ccc(N)cc1)N1CCOCC1)c1ncc(cn1)C(=O)NO |
|---|
| Structure | 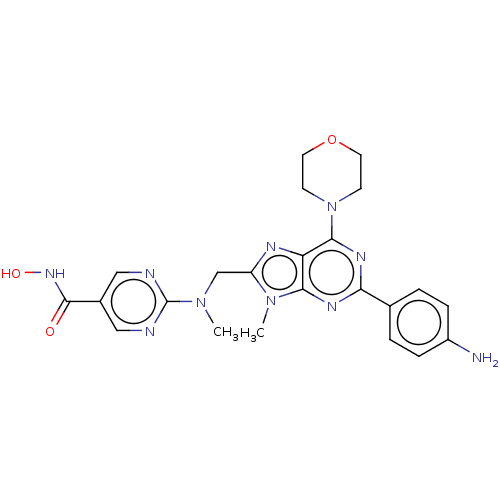
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chen, Y; Wang, X; Xiang, W; He, L; Tang, M; Wang, F; Wang, T; Yang, Z; Yi, Y; Wang, H; Niu, T; Zheng, L; Lei, L; Li, X; Song, H; Chen, L Development of Purine-Based Hydroxamic Acid Derivatives: Potent Histone Deacetylase Inhibitors with Marked in Vitro and in Vivo Antitumor Activities. J Med Chem59:5488-504 (2016) [PubMed] Article
Chen, Y; Wang, X; Xiang, W; He, L; Tang, M; Wang, F; Wang, T; Yang, Z; Yi, Y; Wang, H; Niu, T; Zheng, L; Lei, L; Li, X; Song, H; Chen, L Development of Purine-Based Hydroxamic Acid Derivatives: Potent Histone Deacetylase Inhibitors with Marked in Vitro and in Vivo Antitumor Activities. J Med Chem59:5488-504 (2016) [PubMed] Article