| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
|---|
| Ligand | BDBM50208311 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1632644 (CHEMBL3875436) |
|---|
| IC50 | 117±n/a nM |
|---|
| Citation |  Zhou, ZZ; Ge, BC; Zhong, QP; Huang, C; Cheng, YF; Yang, XM; Wang, HT; Xu, JP Development of highly potent phosphodiesterase 4 inhibitors with anti-neuroinflammation potential: Design, synthesis, and structure-activity relationship study of catecholamides bearing aromatic rings. Eur J Med Chem124:372-379 (2016) [PubMed] Article Zhou, ZZ; Ge, BC; Zhong, QP; Huang, C; Cheng, YF; Yang, XM; Wang, HT; Xu, JP Development of highly potent phosphodiesterase 4 inhibitors with anti-neuroinflammation potential: Design, synthesis, and structure-activity relationship study of catecholamides bearing aromatic rings. Eur J Med Chem124:372-379 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
|---|
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | DPDE2 | PDE46 | PDE4A | PDE4A_HUMAN | Phosphodiesterase 4 (PDE4) | Phosphodiesterase 4A | Phosphodiesterase 4A (PDE4) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 98113.27 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P27815 |
|---|
| Residue: | 886 |
|---|
| Sequence: | MEPPTVPSERSLSLSLPGPREGQATLKPPPQHLWRQPRTPIRIQQRGYSDSAERAERERQ
PHRPIERADAMDTSDRPGLRTTRMSWPSSFHGTGTGSGGAGGGSSRRFEAENGPTPSPGR
SPLDSQASPGLVLHAGAATSQRRESFLYRSDSDYDMSPKTMSRNSSVTSEAHAEDLIVTP
FAQVLASLRSVRSNFSLLTNVPVPSNKRSPLGGPTPVCKATLSEETCQQLARETLEELDW
CLEQLETMQTYRSVSEMASHKFKRMLNRELTHLSEMSRSGNQVSEYISTTFLDKQNEVEI
PSPTMKEREKQQAPRPRPSQPPPPPVPHLQPMSQITGLKKLMHSNSLNNSNIPRFGVKTD
QEELLAQELENLNKWGLNIFCVSDYAGGRSLTCIMYMIFQERDLLKKFRIPVDTMVTYML
TLEDHYHADVAYHNSLHAADVLQSTHVLLATPALDAVFTDLEILAALFAAAIHDVDHPGV
SNQFLINTNSELALMYNDESVLENHHLAVGFKLLQEDNCDIFQNLSKRQRQSLRKMVIDM
VLATDMSKHMTLLADLKTMVETKKVTSSGVLLLDNYSDRIQVLRNMVHCADLSNPTKPLE
LYRQWTDRIMAEFFQQGDRERERGMEISPMCDKHTASVEKSQVGFIDYIVHPLWETWADL
VHPDAQEILDTLEDNRDWYYSAIRQSPSPPPEEESRGPGHPPLPDKFQFELTLEEEEEEE
ISMAQIPCTAQEALTAQGLSGVEEALDATIAWEASPAQESLEVMAQEASLEAELEAVYLT
QQAQSTGSAPVAPDEFSSREEFVVAVSHSSPSALALQSPLLPAWRTLSVSEHAPGLPGLP
STAAEVEAQREHQAAKRACSACAGTFGEDTSALPAPGGGGSGGDPT
|
|
|
|---|
| BDBM50208311 |
|---|
| n/a |
|---|
| Name | BDBM50208311 |
|---|
| Synonyms: | CHEMBL3885161 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H18F2N2O3 |
|---|
| Mol. Mass. | 348.3439 |
|---|
| SMILES | FC(F)Oc1ccc(cc1OC1CCCC1)C(=O)Nc1cccnc1 |
|---|
| Structure | 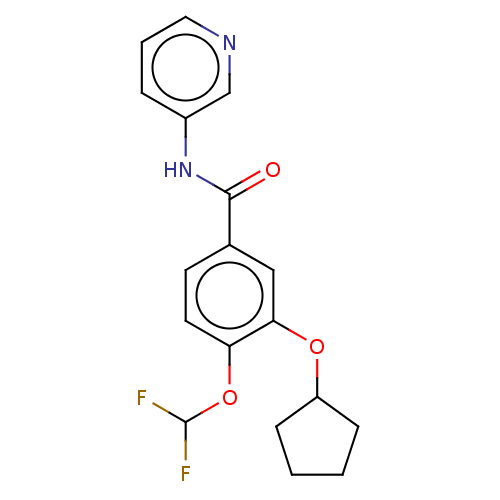
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Zhou, ZZ; Ge, BC; Zhong, QP; Huang, C; Cheng, YF; Yang, XM; Wang, HT; Xu, JP Development of highly potent phosphodiesterase 4 inhibitors with anti-neuroinflammation potential: Design, synthesis, and structure-activity relationship study of catecholamides bearing aromatic rings. Eur J Med Chem124:372-379 (2016) [PubMed] Article
Zhou, ZZ; Ge, BC; Zhong, QP; Huang, C; Cheng, YF; Yang, XM; Wang, HT; Xu, JP Development of highly potent phosphodiesterase 4 inhibitors with anti-neuroinflammation potential: Design, synthesis, and structure-activity relationship study of catecholamides bearing aromatic rings. Eur J Med Chem124:372-379 (2016) [PubMed] Article