| Reaction Details |
|---|
 | Report a problem with these data |
| Target | D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor |
|---|
| Ligand | BDBM50017543 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_60009 (CHEMBL675834) |
|---|
| IC50 | 3.1±n/a nM |
|---|
| Citation |  Anderson, PS; Baldwin, JJ; McClure, DE; Lundell, GF; Jones, JH; Randall, WC; Martin, GE; Williams, M; Hirshfield, JM; Clineschmidt, BV; Lumma, PK; Remy, DC Synthesis of (7R)-7H-indolo[3,4-gh][1,4]benzoxazines, a new class of D-heteroergolines with dopamine agonist activity. J Med Chem26:363-7 (1983) [PubMed] Anderson, PS; Baldwin, JJ; McClure, DE; Lundell, GF; Jones, JH; Randall, WC; Martin, GE; Williams, M; Hirshfield, JM; Clineschmidt, BV; Lumma, PK; Remy, DC Synthesis of (7R)-7H-indolo[3,4-gh][1,4]benzoxazines, a new class of D-heteroergolines with dopamine agonist activity. J Med Chem26:363-7 (1983) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor |
|---|
| Name: | D(1A)/D(1B)/D(2)/D(3)/D(4) dopamine receptor |
|---|
| Synonyms: | Dopamine receptor |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 60018 |
|---|
| Components: | This complex has 5 components. |
|---|
| Component 1 |
| Name: | D(2) dopamine receptor |
|---|
| Synonyms: | DOPAMINE D2 | DOPAMINE D2 Long | DOPAMINE D2 Short | DRD2_RAT | Dopamine D2 receptor | Dopamine2-like | Drd2 |
|---|
| Type: | G Protein-Coupled Receptor (GPCR) |
|---|
| Mol. Mass.: | 50931.60 |
|---|
| Organism: | Rattus norvegicus (rat) |
|---|
| Description: | P61169 |
|---|
| Residue: | 444 |
|---|
| Sequence: | MDPLNLSWYDDDLERQNWSRPFNGSEGKADRPHYNYYAMLLTLLIFIIVFGNVLVCMAVS
REKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTA
SILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMIAIVWVLSFTISCPLLFGLNNTDQN
ECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRKRRKRVNTKRSSRAFRANLKTPL
KGNCTHPEDMKLCTVIMKSNGSFPVNRRRMDAARRAQELEMEMLSSTSPPERTRYSPIPP
SHHQLTLPDPSHHGLHSNPDSPAKPEKNGHAKIVNPRIAKFFEIQTMPNGKTRTSLKTMS
RRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNS
AVNPIIYTTFNIEFRKAFMKILHC
|
|
|
|---|
| Component 2 |
| Name: | D(1A) dopamine receptor |
|---|
| Synonyms: | DOPAMINE D1 | DRD1_RAT | Dopamine D1 high | Dopamine D1 low | Dopamine receptor | Dopamine receptor D1 | Dopamine1-like | Drd1 | Drd1a |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 49429.75 |
|---|
| Organism: | RAT |
|---|
| Description: | P18901 |
|---|
| Residue: | 446 |
|---|
| Sequence: | MAPNTSTMDEAGLPAERDFSFRILTACFLSLLILSTLLGNTLVCAAVIRFRHLRSKVTNF
FVISLAVSDLLVAVLVMPWKAVAEIAGFWPLGPFCNIWVAFDIMCSTASILNLCVISVDR
YWAISSPFQYERKMTPKAAFILISVAWTLSVLISFIPVQLSWHKAKPTWPLDGNFTSLED
TEDDNCDTRLSRTYAISSSLISFYIPVAIMIVTYTSIYRIAQKQIRRISALERAAVHAKN
CQTTAGNGNPVECAQSESSFKMSFKRETKVLKTLSVIMGVFVCCWLPFFISNCMVPFCGS
EETQPFCIDSITFDVFVWFGWANSSLNPIIYAFNADFQKAFSTLLGCYRLCPTTNNAIET
VSINNNGAVVFSSHHEPRGSISKDCNLVYLIPHAVGSSEDLKKEEAGGIAKPLEKLSPAL
SVILDYDTDVSLEKIQPVTHSGQHST
|
|
|
|---|
| Component 3 |
| Name: | D(3) dopamine receptor |
|---|
| Synonyms: | DOPAMINE D3 | DRD3_RAT | Dopamine D3 receptor | Drd3 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 49540.58 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P19020 |
|---|
| Residue: | 446 |
|---|
| Sequence: | MAPLSQISTHLNSTCGAENSTGVNRARPHAYYALSYCALILAIIFGNGLVCAAVLRERAL
QTTTNYLVVSLAVADLLVATLVMPWVVYLEVTGGVWNFSRICCDVFVTLDVMMCTASILN
LCAISIDRYTAVVMPVHYQHGTGQSSCRRVALMITAVWVLAFAVSCPLLFGFNTTGDPSI
CSISNPDFVIYSSVVSFYVPFGVTVLVYARIYIVLRQRQRKRILTRQNSQCISIRPGFPQ
QSSCLRLHPIRQFSIRARFLSDATGQMEHIEDKQYPQKCQDPLLSHLQPPSPGQTHGGLK
RYYSICQDTALRHPSLEGGAGMSPVERTRNSLSPTMAPKLSLEVRKLSNGRLSTSLRLGP
LQPRGVPLREKKATQMVVIVLGAFIVCWLPFFLTHVLNTHCQACHVSPELYRATTWLGYV
NSALNPVIYTTFNVEFRKAFLKILSC
|
|
|
|---|
| Component 4 |
| Name: | D(4) dopamine receptor |
|---|
| Synonyms: | D(4) dopamine receptor | DOPAMINE D4 | DOPAMINE D4.4 | DRD4_RAT | Dopamine receptor | Drd4 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 41307.65 |
|---|
| Organism: | RAT |
|---|
| Description: | DOPAMINE D4.4 0 RAT::P30729 |
|---|
| Residue: | 387 |
|---|
| Sequence: | MGNSSATGDGGLLAGRGPESLGTGTGLGGAGAAALVGGVLLIGMVLAGNSLVCVSVASER
ILQTPTNYFIVSLAAADLLLAVLVLPLFVYSEVQGGVWLLSPRLCDTLMAMDVMLCTASI
FNLCAISVDRFVAVTVPLRYNQQGQCQLLLIAATWLLSAAVAAPVVCGLNDVPGRDPTVC
CLEDRDYVVYSSICSFFLPCPLMLLLYWATFRGLRRWEAARHTKLHSRAPRRPSGPGPPV
SDPTQGPLFSDCPPPSPSLRTSPTVSSRPESDLSQSPCSPGCLLPDAALAQPPAPSSRRK
RGAKITGRERKAMRVLPVVVGAFLMCWTPFFVVHITRALCPACFVSPRLVSAVTWLGYVN
SALNPIIYTIFNAEFRSVFRKTLRLRC
|
|
|
|---|
| Component 5 |
| Name: | D(1B) dopamine receptor |
|---|
| Synonyms: | D(1B) dopamine receptor | DOPAMINE D5 | DRD5_RAT | Dopamine D1B | Dopamine D5 receptor | Dopamine receptor | Drd5 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 52827.88 |
|---|
| Organism: | RAT |
|---|
| Description: | Dopamine D1B 0 RAT::P25115 |
|---|
| Residue: | 475 |
|---|
| Sequence: | MLPPGRNRTAQPARLGLQRQLAQVDAPAGSATPLGPAQVVTAGLLTLLIVWTLLGNVLVC
AAIVRSRHLRAKMTNIFIVSLAVSDLFVALLVMPWKAVAEVAGYWPFGTFCDIWVAFDIM
CSTASILNLCIISVDRYWAISRPFRYERKMTQRVALVMVGLAWTLSILISFIPVQLNWHR
DKAGSQGQEGLLSNGTPWEEGWELEGRTENCDSSLNRTYAISSSLISFYIPVAIMIVTYT
RIYRIAQVQIRRISSLERAAEHAQSCRSRGAYEPDPSLRASIKKETKVFKTLSMIMGVFV
CCWLPFFILNCMVPFCSSGDAEGPKTGFPCVSETTFDIFVWFGWANSSLNPIIYAFNADF
RKVFAQLLGCSHFCFRTPVQTVNISNELISYNQDTVFHKEIATAYVHMIPNAVSSGDREV
GEEEEEGPFDHMSQISPTTPDGDLAAESVWELDCEEEVSLGKISPLTPNCFDKTA
|
|
|
|---|
| BDBM50017543 |
|---|
| n/a |
|---|
| Name | BDBM50017543 |
|---|
| Synonyms: | (6aR,9R,10aR)-9-Methylsulfanylmethyl-7-propyl-4,6,6a,7,8,9,10,10a-octahydro-indolo[4,3-fg]quinolin-7-ium | (6aR,9R,10aR)-9-Methylsulfanylmethyl-7-propyl-4,6,6a,7,8,9,10,10a-octahydro-indolo[4,3-fg]quinoline | 5-Bromo-7-methyl-4,6,6a,7,8,9-hexahydro-indolo[4,3-fg]quinoline-9-carboxylic acid (10b-hydroxy-5-isobutyl-2-isopropyl-3,6-dioxo-octahydro-oxazolo[3,2-a]pyrrolo[2,1-c]pyrazin-2-yl)-amide | 9-Methylsulfanylmethyl-7-propyl-4,6,6a,7,8,9,10,10a-octahydro-indolo[4,3-fg]quinolin-7-ium(Pergolide) | 9-Methylsulfanylmethyl-7-propyl-4,6,6a,7,8,9,10,10a-octahydro-indolo[4,3-fg]quinoline | 9-Methylsulfanylmethyl-7-propyl-4,6,6a,7,8,9,10,10a-octahydro-indolo[4,3-fg]quinoline(pergolide) | 9-Methylsulfanylmethyl-7-propyl-4,6,6a,7,8,9,10,10a-octahydro-indolo[4,3-fg]quinoline, mesylate (Pergolide) | CHEMBL531 | PERGOLIDE | Permax | [2-(1H-Indol-4-yl)-ethyl]-methyl-amine | cid_47811 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H26N2S |
|---|
| Mol. Mass. | 314.488 |
|---|
| SMILES | CCCN1C[C@H](CSC)C[C@H]2[C@H]1Cc1c[nH]c3cccc2c13 |r| |
|---|
| Structure | 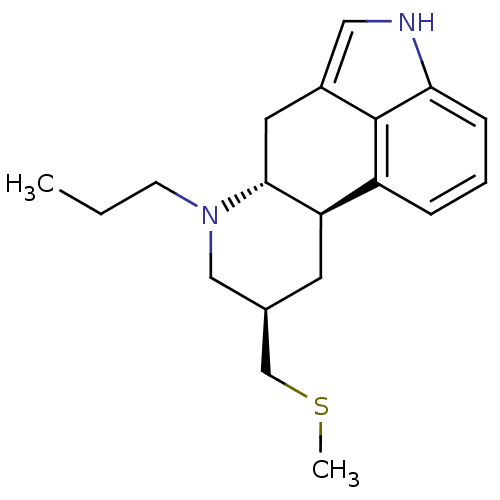
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Anderson, PS; Baldwin, JJ; McClure, DE; Lundell, GF; Jones, JH; Randall, WC; Martin, GE; Williams, M; Hirshfield, JM; Clineschmidt, BV; Lumma, PK; Remy, DC Synthesis of (7R)-7H-indolo[3,4-gh][1,4]benzoxazines, a new class of D-heteroergolines with dopamine agonist activity. J Med Chem26:363-7 (1983) [PubMed]
Anderson, PS; Baldwin, JJ; McClure, DE; Lundell, GF; Jones, JH; Randall, WC; Martin, GE; Williams, M; Hirshfield, JM; Clineschmidt, BV; Lumma, PK; Remy, DC Synthesis of (7R)-7H-indolo[3,4-gh][1,4]benzoxazines, a new class of D-heteroergolines with dopamine agonist activity. J Med Chem26:363-7 (1983) [PubMed]