| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lactoylglutathione lyase |
|---|
| Ligand | BDBM7461 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_71295 (CHEMBL684310) |
|---|
| Ki | 83200±n/a nM |
|---|
| Citation |  Doweyko, AM The hypothetical active site lattice. An approach to modelling active sites from data on inhibitor molecules. J Med Chem31:1396-406 (1988) [PubMed] Doweyko, AM The hypothetical active site lattice. An approach to modelling active sites from data on inhibitor molecules. J Med Chem31:1396-406 (1988) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lactoylglutathione lyase |
|---|
| Name: | Lactoylglutathione lyase |
|---|
| Synonyms: | Aldoketomutase | GLO1 | Glx I | Glyoxalase 1 (GLO1) | Glyoxalase I | Ketone-aldehyde mutase | LGUL_HUMAN | Methylglyoxalase | S-D-lactoylglutathione methylglyoxal lyase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 20772.95 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q04760 |
|---|
| Residue: | 184 |
|---|
| Sequence: | MAEPQPPSGGLTDEAALSCCSDADPSTKDFLLQQTMLRVKDPKKSLDFYTRVLGMTLIQK
CDFPIMKFSLYFLAYEDKNDIPKEKDEKIAWALSRKATLELTHNWGTEDDETQSYHNGNS
DPRGFGHIGIAVPDVYSACKRFEELGVKFVKKPDDGKMKGLAFIQDPDGYWIEILNPNKM
ATLM
|
|
|
|---|
| BDBM7461 |
|---|
| n/a |
|---|
| Name | BDBM7461 |
|---|
| Synonyms: | 5,7-dihydroxy-2-phenyl-4H-chromen-4-one | 5,7-dihydroxy-2-phenyl-chromen-4-one | 5,7-dihydroxyflavone | CHEMBL117 | chrysin | cid_5281607 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H10O4 |
|---|
| Mol. Mass. | 254.2375 |
|---|
| SMILES | Oc1cc(O)c2c(c1)oc(cc2=O)-c1ccccc1 |
|---|
| Structure | 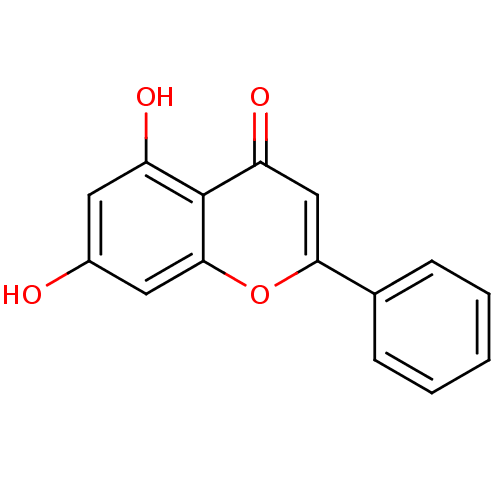
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Doweyko, AM The hypothetical active site lattice. An approach to modelling active sites from data on inhibitor molecules. J Med Chem31:1396-406 (1988) [PubMed]
Doweyko, AM The hypothetical active site lattice. An approach to modelling active sites from data on inhibitor molecules. J Med Chem31:1396-406 (1988) [PubMed]