| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 5-hydroxytryptamine receptor 3A/3B |
|---|
| Ligand | BDBM50000492 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_3109 (CHEMBL620587) |
|---|
| Ki | 1.3±n/a nM |
|---|
| Citation |  Rosen, T; Seeger, TF; McLean, S; Nagel, AA; Ives, JL; Guarino, KJ; Bryce, D; Furman, J; Roth, RW; Chalabi, PM Synthesis, in vitro binding profile, and central nervous system penetrability of the highly potent 5-HT3 receptor antagonist [3H]-4-(2-methoxyphenyl)-2-[4(5)-methyl-5(4)-imidazolylmethyl]thiazole. J Med Chem33:3020-3 (1990) [PubMed] Rosen, T; Seeger, TF; McLean, S; Nagel, AA; Ives, JL; Guarino, KJ; Bryce, D; Furman, J; Roth, RW; Chalabi, PM Synthesis, in vitro binding profile, and central nervous system penetrability of the highly potent 5-HT3 receptor antagonist [3H]-4-(2-methoxyphenyl)-2-[4(5)-methyl-5(4)-imidazolylmethyl]thiazole. J Med Chem33:3020-3 (1990) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 5-hydroxytryptamine receptor 3A/3B |
|---|
| Name: | 5-hydroxytryptamine receptor 3A/3B |
|---|
| Synonyms: | Serotonin 3 receptor (5HT3) |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 357762 |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | 5-hydroxytryptamine receptor 3A |
|---|
| Synonyms: | 5-HT-3 | 5-HT3-A | 5-HT3A | 5-HT3A Serotonin Receptor | 5-HT3R | 5-hydroxytryptamine receptor 3 | 5-hydroxytryptamine receptor 3A | 5HT3A_MOUSE | 5ht3 | Htr3 | Htr3a | Serotonin 3 receptor (5HT3) | Serotonin 3a (5-HT3a) receptor | Serotonin receptor 3A | Serotonin-gated ion channel receptor |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 56056.00 |
|---|
| Organism: | Mus musculus (house mouse) |
|---|
| Description: | 5HT3A |
|---|
| Residue: | 487 |
|---|
| Sequence: | MRLCIPQVLLALFLSMLTAPGEGSRRRATQEDTTQPALLRLSDHLLANYKKGVRPVRDWR
KPTTVSIDVIMYAILNVDEKNQVLTTYIWYRQYWTDEFLQWTPEDFDNVTKLSIPTDSIW
VPDILINEFVDVGKSPNIPYVYVHHRGEVQNYKPLQLVTACSLDIYNFPFDVQNCSLTFT
SWLHTIQDINITLWRSPEEVRSDKSIFINQGEWELLEVFPQFKEFSIDISNSYAEMKFYV
IIRRRPLFYAVSLLLPSIFLMVVDIVGFCLPPDSGERVSFKITLLLGYSVFLIIVSDTLP
ATIGTPLIGVYFVVCMALLVISLAETIFIVRLVHKQDLQRPVPDWLRHLVLDRIAWILCL
GEQPMAHRPPATFQANKTDDCSGSDLLPAMGNHCSHVGGPQDLEKTPRGRGSPLPPPREA
SLAVRGLLQELSSIRHFLEKRDEMREVARDWLRVGYVLDRLLFRIYLLAVLAYSITLVTL
WSIWHYS
|
|
|
|---|
| Component 2 |
| Name: | 5-hydroxytryptamine receptor 3B |
|---|
| Synonyms: | 5HT3B_MOUSE | Htr3b | Serotonin 3 receptor (5HT3) | Serotonin 3b (5-HT3b) receptor |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 50317.35 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_3121 |
|---|
| Residue: | 437 |
|---|
| Sequence: | MILLWSCLLVAVVGILGTATPQPGNSSLHRLTRQLLQQYHKEVRPVYNWAEATTVYLDLC
VHAVLDVDVQNQKLKTSVWYREVWNDEFLSWNSSLFDEIQEISLPLSALWAPDIIINEFV
DVERSPDLPYVYVNSSGTIRNHKPIQVVSACSLQTYAFPFDIQNCSLTFNSILHTVEDID
LGFLRNREDIENDKRAFMNDSEWQLLSVSSTYHIRQSSAGDFAQIRFNVVIRRCPLAYVV
SLLIPSIFLMLVDLGSFYLPPNCRARIVFKTNVLVGYTVFRVNMSDEVPRSAGCTPLIGV
FFTVCMALLVLSLSKSILLIKFLYEERHSGQERPLMCLQGDSDAEESRLYLGAPRADVTE
SPVHQEHRVPSDTLKDFWFQFRSINNSLRTRDQIHQKEVEWLAILYRFDQLLFRIYLAVL
GLYTVTLCSLWALWSRM
|
|
|
|---|
| BDBM50000492 |
|---|
| n/a |
|---|
| Name | BDBM50000492 |
|---|
| Synonyms: | (zacopride)4-Amino-N-(1-aza-bicyclo[2.2.2]oct-3-yl)-5-chloro-2-methoxy-benzamide | 4-Amino-N-(1-aza-bicyclo[2.2.2]oct-3-yl)-5-chloro-2-methoxy-benzamide | 4-Amino-N-(1-aza-bicyclo[2.2.2]oct-3-yl)-5-chloro-2-methoxy-benzamide (Zacopride) | 4-Amino-N-(1-aza-bicyclo[2.2.2]oct-3-yl)-5-chloro-2-methoxy-benzamide(zacopride) | CHEMBL18041 | HTR5A4-Amino-N-(1-aza-bicyclo[2.2.2]oct-3-yl)-5-chloro-2-methoxy-benzamide | ZACOPRIDE | ZACOPRIDE,R | ZACOPRIDE,RS | ZACOPRIDE,S |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H20ClN3O2 |
|---|
| Mol. Mass. | 309.791 |
|---|
| SMILES | COc1cc(N)c(Cl)cc1C(=O)NC1CN2CCC1CC2 |(27.19,-33.96,;28.52,-34.73,;28.53,-36.27,;27.2,-37.04,;27.2,-38.58,;25.86,-39.35,;28.53,-39.36,;28.53,-40.89,;29.87,-38.58,;29.86,-37.03,;31.19,-36.26,;31.19,-34.72,;32.53,-37.02,;33.86,-36.25,;35.2,-37.02,;36.52,-36.25,;36.52,-34.71,;35.19,-33.94,;33.85,-34.71,;34.61,-36.04,;35.74,-34.91,)| |
|---|
| Structure | 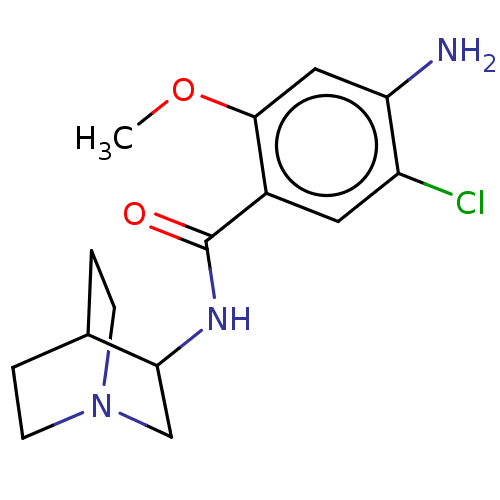
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Rosen, T; Seeger, TF; McLean, S; Nagel, AA; Ives, JL; Guarino, KJ; Bryce, D; Furman, J; Roth, RW; Chalabi, PM Synthesis, in vitro binding profile, and central nervous system penetrability of the highly potent 5-HT3 receptor antagonist [3H]-4-(2-methoxyphenyl)-2-[4(5)-methyl-5(4)-imidazolylmethyl]thiazole. J Med Chem33:3020-3 (1990) [PubMed]
Rosen, T; Seeger, TF; McLean, S; Nagel, AA; Ives, JL; Guarino, KJ; Bryce, D; Furman, J; Roth, RW; Chalabi, PM Synthesis, in vitro binding profile, and central nervous system penetrability of the highly potent 5-HT3 receptor antagonist [3H]-4-(2-methoxyphenyl)-2-[4(5)-methyl-5(4)-imidazolylmethyl]thiazole. J Med Chem33:3020-3 (1990) [PubMed]