| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Alpha-2A/Alpha-2B/Alpha-2C adrenergic receptor |
|---|
| Ligand | BDBM50001922 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_33314 (CHEMBL645610) |
|---|
| IC50 | 0.001100±n/a nM |
|---|
| Citation |  Amemiya, Y; Hong, SS; Venkataraman, BV; Patil, PN; Shams, G; Romstedt, K; Feller, DR; Hsu, FL; Miller, DD Synthesis and alpha-adrenergic activities of 2- and 4-substituted imidazoline and imidazole analogues. J Med Chem35:750-5 (1992) [PubMed] Amemiya, Y; Hong, SS; Venkataraman, BV; Patil, PN; Shams, G; Romstedt, K; Feller, DR; Hsu, FL; Miller, DD Synthesis and alpha-adrenergic activities of 2- and 4-substituted imidazoline and imidazole analogues. J Med Chem35:750-5 (1992) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Alpha-2A/Alpha-2B/Alpha-2C adrenergic receptor |
|---|
| Name: | Alpha-2A/Alpha-2B/Alpha-2C adrenergic receptor |
|---|
| Synonyms: | Adrenergic Receptors (alpha 2) | Adrenergic receptor alpha-2 |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 305672 |
|---|
| Components: | This complex has 3 components. |
|---|
| Component 1 |
| Name: | Alpha-2B adrenergic receptor |
|---|
| Synonyms: | ADA2B_HUMAN | ADRA2B | ADRA2L1 | ADRA2RL1 | Adrenergic alpha2B | Adrenergic receptor | Adrenergic receptor alpha | Adrenergic, alpha-2B-, receptor [Homo sapiens] | Alpha-2 adrenergic receptor subtype C2 | Alpha-2B adrenoceptor | Alpha-2B adrenoreceptor | Alpha-2BAR |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 49964.20 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P18089 |
|---|
| Residue: | 450 |
|---|
| Sequence: | MDHQDPYSVQATAAIAAAITFLILFTIFGNALVILAVLTSRSLRAPQNLFLVSLAAADIL
VATLIIPFSLANELLGYWYFRRTWCEVYLALDVLFCTSSIVHLCAISLDRYWAVSRALEY
NSKRTPRRIKCIILTVWLIAAVISLPPLIYKGDQGPQPRGRPQCKLNQEAWYILASSIGS
FFAPCLIMILVYLRIYLIAKRSNRRGPRAKGGPGQGESKQPRPDHGGALASAKLPALASV
ASAREVNGHSKSTGEKEEGETPEDTGTRALPPSWAALPNSGQGQKEGVCGASPEDEAEEE
EEEEEEEEECEPQAVPVSPASACSPPLQQPQGSRVLATLRGQVLLGRGVGAIGGQWWRRR
AQLTREKRFTFVLAVVIGVFVLCWFPFFFSYSLGAICPKHCKVPHGLFQFFFWIGYCNSS
LNPVIYTIFNQDFRRAFRRILCRPWTQTAW
|
|
|
|---|
| Component 2 |
| Name: | Alpha-2A adrenergic receptor |
|---|
| Synonyms: | ADA2A_HUMAN | ADRA2A | ADRA2R | ADRAR | Adrenergic alpha2A | Adrenergic receptor alpha | Alpha-2 adrenergic receptor subtype C10 | Alpha-2A adrenoceptor | Alpha-2A adrenoreceptor | Alpha-2AAR | alpha-2A adrenergic receptor [Homo sapiens] |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48979.91 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P08913 |
|---|
| Residue: | 450 |
|---|
| Sequence: | MGSLQPDAGNASWNGTEAPGGGARATPYSLQVTLTLVCLAGLLMLLTVFGNVLVIIAVFT
SRALKAPQNLFLVSLASADILVATLVIPFSLANEVMGYWYFGKAWCEIYLALDVLFCTSS
IVHLCAISLDRYWSITQAIEYNLKRTPRRIKAIIITVWVISAVISFPPLISIEKKGGGGG
PQPAEPRCEINDQKWYVISSCIGSFFAPCLIMILVYVRIYQIAKRRTRVPPSRRGPDAVA
APPGGTERRPNGLGPERSAGPGGAEAEPLPTQLNGAPGEPAPAGPRDTDALDLEESSSSD
HAERPPGPRRPERGPRGKGKARASQVKPGDSLPRRGPGATGIGTPAAGPGEERVGAAKAS
RWRGRQNREKRFTFVLAVVIGVFVVCWFPFFFTYTLTAVGCSVPRTLFKFFFWFGYCNSS
LNPVIYTIFNHDFRRAFKKILCRGDRKRIV
|
|
|
|---|
| Component 3 |
| Name: | Alpha-2C adrenergic receptor |
|---|
| Synonyms: | ADA2C_HUMAN | ADRA2C | ADRA2L2 | ADRA2RL2 | Adrenergic alpha2C | Adrenergic receptor | Adrenergic receptor alpha | Adrenergic, alpha-2C-, receptor | Alpha-2 adrenergic receptor subtype C4 | Alpha-2C adrenoceptor | Alpha-2C adrenoreceptor | adrenergic, alpha-2C-, receptor [Homo sapiens] |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 49552.32 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P18825 |
|---|
| Residue: | 462 |
|---|
| Sequence: | MASPALAAALAVAAAAGPNASGAGERGSGGVANASGASWGPPRGQYSAGAVAGLAAVVGF
LIVFTVVGNVLVVIAVLTSRALRAPQNLFLVSLASADILVATLVMPFSLANELMAYWYFG
QVWCGVYLALDVLFCTSSIVHLCAISLDRYWSVTQAVEYNLKRTPRRVKATIVAVWLISA
VISFPPLVSLYRQPDGAAYPQCGLNDETWYILSSCIGSFFAPCLIMGLVYARIYRVAKLR
TRTLSEKRAPVGPDGASPTTENGLGAAAGAGENGHCAPPPADVEPDESSAAAERRRRRGA
LRRGGRRRAGAEGGAGGADGQGAGPGAAESGALTASRSPGPGGRLSRASSRSVEFFLSRR
RRARSSVCRRKVAQAREKRFTFVLAVVMGVFVLCWFPFFFSYSLYGICREACQVPGPLFK
FFFWIGYCNSSLNPVIYTVFNQDFRRSFKHILFRRRRRGFRQ
|
|
|
|---|
| BDBM50001922 |
|---|
| n/a |
|---|
| Name | BDBM50001922 |
|---|
| Synonyms: | 2-Naphthalen-1-ylmethyl-4,5-dihydro-1H-imidazole; hydrochloride | Albalon | CHEMBL1706 | NAPHAZOLINE HYDROCHLORIDE | Nafazair | Naphazoline | Naphazoline Nitrate | Naphcon forte | Opcon | Vasocon | cid_11079 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H14N2 |
|---|
| Mol. Mass. | 210.2744 |
|---|
| SMILES | C(C1=NCCN1)c1cccc2ccccc12 |t:1| |
|---|
| Structure | 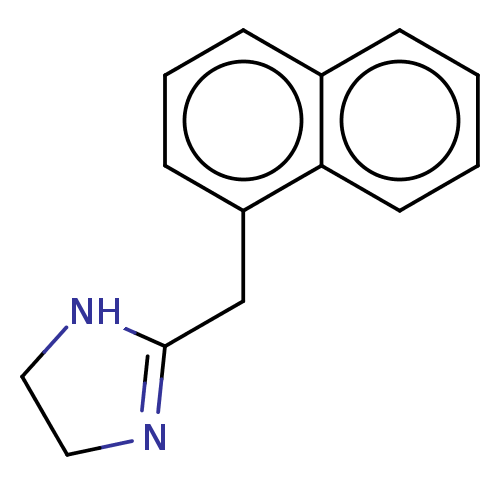
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Amemiya, Y; Hong, SS; Venkataraman, BV; Patil, PN; Shams, G; Romstedt, K; Feller, DR; Hsu, FL; Miller, DD Synthesis and alpha-adrenergic activities of 2- and 4-substituted imidazoline and imidazole analogues. J Med Chem35:750-5 (1992) [PubMed]
Amemiya, Y; Hong, SS; Venkataraman, BV; Patil, PN; Shams, G; Romstedt, K; Feller, DR; Hsu, FL; Miller, DD Synthesis and alpha-adrenergic activities of 2- and 4-substituted imidazoline and imidazole analogues. J Med Chem35:750-5 (1992) [PubMed]