| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Protein mono-ADP-ribosyltransferase PARP4 |
|---|
| Ligand | BDBM27135 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_1646307 |
|---|
| IC50 | 3467±n/a nM |
|---|
| Citation |  Thorsell, AG; Ekblad, T; Karlberg, T; Löw, M; Pinto, AF; Trésaugues, L; Moche, M; Cohen, MS; Schüler, H Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors. J Med Chem60:1262-1271 (2017) [PubMed] Article Thorsell, AG; Ekblad, T; Karlberg, T; Löw, M; Pinto, AF; Trésaugues, L; Moche, M; Cohen, MS; Schüler, H Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors. J Med Chem60:1262-1271 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Protein mono-ADP-ribosyltransferase PARP4 |
|---|
| Name: | Protein mono-ADP-ribosyltransferase PARP4 |
|---|
| Synonyms: | (ARTD4 or PARP4) | 193 kDa vault protein | 2.4.2.- | 2.4.2.30 | ADP-ribosyltransferase diphtheria toxin-like 4 | ADPRTL1 | ADPRTL1 | ARTD4 | KIAA0177 | KIAA0177 GN | PARP-4 | PARP-related/IalphaI-related H5/proline-rich | PARP4 | PARP4_HUMAN | PARPL | PH5P | Poly [ADP-ribose] polymerase 4 | Synonyms=ADPRTL1 | VPARP | Vault poly(ADP-ribose) polymerase |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 192563.23 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9UKK3 |
|---|
| Residue: | 1724 |
|---|
| Sequence: | MVMGIFANCIFCLKVKYLPQQQKKKLQTDIKENGGKFSFSLNPQCTHIILDNADVLSQYQ
LNSIQKNHVHIANPDFIWKSIREKRLLDVKNYDPYKPLDITPPPDQKASSSEVKTEGLCP
DSATEEEDTVELTEFGMQNVEIPHLPQDFEVAKYNTLEKVGMEGGQEAVVVELQCSRDSR
DCPFLISSHFLLDDGMETRRQFAIKKTSEDASEYFENYIEELKKQGFLLREHFTPEATQL
ASEQLQALLLEEVMNSSTLSQEVSDLVEMIWAEALGHLEHMLLKPVNRISLNDVSKAEGI
LLLVKAALKNGETAEQLQKMMTEFYRLIPHKGTMPKEVNLGLLAKKADLCQLIRDMVNVC
ETNLSKPNPPSLAKYRALRCKIEHVEQNTEEFLRVRKEVLQNHHSKSPVDVLQIFRVGRV
NETTEFLSKLGNVRPLLHGSPVQNIVGILCRGLLLPKVVEDRGVQRTDVGNLGSGIYFSD
SLSTSIKYSHPGETDGTRLLLICDVALGKCMDLHEKDFSLTEAPPGYDSVHGVSQTASVT
TDFEDDEFVVYKTNQVKMKYIIKFSMPGDQIKDFHPSDHTELEEYRPEFSNFSKVEDYQL
PDAKTSSSTKAGLQDASGNLVPLEDVHIKGRIIDTVAQVIVFQTYTNKSHVPIEAKYIFP
LDDKAAVCGFEAFINGKHIVGEIKEKEEAQQEYLEAVTQGHGAYLMSQDAPDVFTVSVGN
LPPKAKVLIKITYITELSILGTVGVFFMPATVAPWQQDKALNENLQDTVEKICIKEIGTK
QSFSLTMSIEMPYVIEFIFSDTHELKQKRTDCKAVISTMEGSSLDSSGFSLHIGLSAAYL
PRMWVEKHPEKESEACMLVFQPDLDVDLPDLASESEVIICLDCSSSMEGVTFLQAKQIAL
HALSLVGEKQKVNIIQFGTGYKELFSYPKHITSNTMAAEFIMSATPTMGNTDFWKTLRYL
SLLYPARGSRNILLVSDGHLQDESLTLQLVKRSRPHTRLFACGIGSTANRHVLRILSQCG
AGVFEYFNAKSKHSWRKQIEDQMTRLCSPSCHSVSVKWQQLNPDVPEALQAPAQVPSLFL
NDRLLVYGFIPHCTQATLCALIQEKEFRTMVSTTELQKTTGTMIHKLAARALIRDYEDGI
LHENETSHEMKKQTLKSLIIKLSKENSLITQFTSFVAVEKRDENESPFPDIPKVSELIAK
EDVDFLPYMSWQGEPQEAVRNQSLLASSEWPELRLSKRKHRKIPFSKRKMELSQPEVSED
FEEDGLGVLPAFTSNLERGGVEKLLDLSWTESCKPTATEPLFKKVSPWETSTSSFFPILA
PAVGSYLPPTARAHSPASLSFASYRQVASFGSAAPPRQFDASQFSQGPVPGTCADWIPQS
ASCPTGPPQNPPSSPYCGIVFSGSSLSSAQSAPLQHPGGFTTRPSAGTFPELDSPQLHFS
LPTDPDPIRGFGSYHPSASSPFHFQPSAASLTANLRLPMASALPEALCSQSRTTPVDLCL
LEESVGSLEGSRCPVFAFQSSDTESDELSEVLQDSCFLQIKCDTKDDSILCFLEVKEEDE
IVCIQHWQDAVPWTELLSLQTEDGFWKLTPELGLILNLNTNGLHSFLKQKGIQSLGVKGR
ECLLDLIATMLVLQFIRTRLEKEGIVFKSLMKMDDASISRNIPWAFEAIKQASEWVRRTE
GQYPSICPRLELGNDWDSATKQLLGLQPISTVSPLHRVLHYSQG
|
|
|
|---|
| BDBM27135 |
|---|
| n/a |
|---|
| Name | BDBM27135 |
|---|
| Synonyms: | 2-[(2R)-2-methylpyrrolidin-2-yl]-1H-1,3-benzodiazole-4-carboxamide | VELIPARIB | benzimidazole carboxamide, 3a |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H16N4O |
|---|
| Mol. Mass. | 244.2923 |
|---|
| SMILES | C[C@@]1(CCCN1)c1nc2cccc(C(N)=O)c2[nH]1 |r| |
|---|
| Structure | 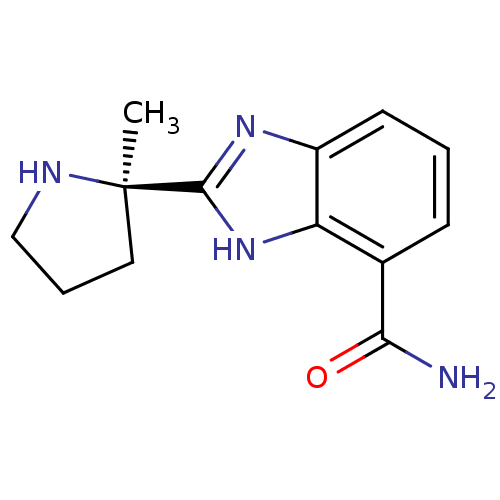
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Thorsell, AG; Ekblad, T; Karlberg, T; Löw, M; Pinto, AF; Trésaugues, L; Moche, M; Cohen, MS; Schüler, H Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors. J Med Chem60:1262-1271 (2017) [PubMed] Article
Thorsell, AG; Ekblad, T; Karlberg, T; Löw, M; Pinto, AF; Trésaugues, L; Moche, M; Cohen, MS; Schüler, H Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors. J Med Chem60:1262-1271 (2017) [PubMed] Article