| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Ligand | BDBM50233393 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1650773 (CHEMBL3999907) |
|---|
| IC50 | 8000±n/a nM |
|---|
| Citation |  Gao, X; Wang, J; Liu, J; Guiadeen, D; Krikorian, A; Boga, SB; Alhassan, AB; Selyutin, O; Yu, W; Yu, Y; Anand, R; Liu, S; Yang, C; Wu, H; Cai, J; Cooper, A; Zhu, H; Maloney, K; Gao, YD; Fischmann, TO; Presland, J; Mansueto, M; Xu, Z; Leccese, E; Zhang-Hoover, J; Knemeyer, I; Garlisi, CG; Bays, N; Stivers, P; Brandish, PE; Hicks, A; Kim, R; Kozlowski, JA Discovery of novel BTK inhibitors with carboxylic acids. Bioorg Med Chem Lett27:1471-1477 (2017) [PubMed] Article Gao, X; Wang, J; Liu, J; Guiadeen, D; Krikorian, A; Boga, SB; Alhassan, AB; Selyutin, O; Yu, W; Yu, Y; Anand, R; Liu, S; Yang, C; Wu, H; Cai, J; Cooper, A; Zhu, H; Maloney, K; Gao, YD; Fischmann, TO; Presland, J; Mansueto, M; Xu, Z; Leccese, E; Zhang-Hoover, J; Knemeyer, I; Garlisi, CG; Bays, N; Stivers, P; Brandish, PE; Hicks, A; Kim, R; Kozlowski, JA Discovery of novel BTK inhibitors with carboxylic acids. Bioorg Med Chem Lett27:1471-1477 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Name: | Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Synonyms: | CAC1C_HUMAN | CACH2 | CACN2 | CACNA1C | CACNL1A1 | CCHL1A1 | Calcium channel (Type L) | Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle | L-type calcium channel alpha-1c/beta-2/alpha2delta-1 | Voltage-dependent L-type calcium channel subunit alpha-1C | Voltage-gated L-type calcium channel | Voltage-gated L-type calcium channel alpha-1C subunit | Voltage-gated calcium channel | Voltage-gated calcium channel subunit alpha Cav1.2 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 248979.79 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Calcium channel (Type L) 0 HUMAN::Q13936 |
|---|
| Residue: | 2221 |
|---|
| Sequence: | MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAAR
QAKLMGSAGNATISTVSSTQRKRQQYGKPKKQGSTTATRPPRALLCLTLKNPIRRACISI
VEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA
YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFR
VLRPLRLVSGVPSLQVVLNSIIKAMVPLLHIALLVLFVIIIYAIIGLELFMGKMHKTCYN
QEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI
TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGD
FQKLREKQQLEEDLKGYLDWITQAEDIDPENEDEGMDEEKPRNMSMPTSETESVNTENVA
GGDIEGENCGARLAHRISKSKFSRYWRRWNRFCRRKCRAAVKSNVFYWLVIFLVFLNTLT
IASEHYNQPNWLTEVQDTANKALLALFTAEMLLKMYSLGLQAYFVSLFNRFDCFVVCGGI
LETILVETKIMSPLGISVLRCVRLLRIFKITRYWNSLSNLVASLLNSVRSIASLLLLLFL
FIIIFSLLGMQLFGGKFNFDEMQTRRSTFDNFPQSLLTVFQILTGEDWNSVMYDGIMAYG
GPSFPGMLVCIYFIILFICGNYILLNVFLAIAVDNLADAESLTSAQKEEEEEKERKKLAR
TASPEKKQELVEKPAVGESKEEKIELKSITADGESPPATKINMDDLQPNENEDKSPYPNP
ETTGEEDEEEPEMPVGPRPRPLSELHLKEKAVPMPEASAFFIFSSNNRFRLQCHRIVNDT
IFTNLILFFILLSSISLAAEDPVQHTSFRNHILFYFDIVFTTIFTIEIALKILGNADYVF
TSIFTLEIILKMTAYGAFLHKGSFCRNYFNILDLLVVSVSLISFGIQSSAINVVKILRVL
RVLRPLRAINRAKGLKHVVQCVFVAIRTIGNIVIVTTLLQFMFACIGVQLFKGKLYTCSD
SSKQTEAECKGNYITYKDGEVDHPIIQPRSWENSKFDFDNVLAAMMALFTVSTFEGWPEL
LYRSIDSHTEDKGPIYNYRVEISIFFIIYIIIIAFFMMNIFVGFVIVTFQEQGEQEYKNC
ELDKNQRQCVEYALKARPLRRYIPKNQHQYKVWYVVNSTYFEYLMFVLILLNTICLAMQH
YGQSCLFKIAMNILNMLFTGLFTVEMILKLIAFKPKGYFSDPWNVFDFLIVIGSIIDVIL
SETNHYFCDAWNTFDALIVVGSIVDIAITEVNPAEHTQCSPSMNAEENSRISITFFRLFR
VMRLVKLLSRGEGIRTLLWTFIKSFQALPYVALLIVMLFFIYAVIGMQVFGKIALNDTTE
INRNNNFQTFPQAVLLLFRCATGEAWQDIMLACMPGKKCAPESEPSNSTEGETPCGSSFA
VFYFISFYMLCAFLIINLFVAVIMDNFDYLTRDWSILGPHHLDEFKRIWAEYDPEAKGRI
KHLDVVTLLRRIQPPLGFGKLCPHRVACKRLVSMNMPLNSDGTVMFNATLFALVRTALRI
KTEGNLEQANEELRAIIKKIWKRTSMKLLDQVVPPAGDDEVTVGKFYATFLIQEYFRKFK
KRKEQGLVGKPSQRNALSLQAGLRTLHDIGPEIRRAISGDLTAEEELDKAMKEAVSAASE
DDIFRRAGGLFGNHVSYYQSDGRSAFPQTFTTQRPLHINKAGSSQGDTESPSHEKLVDST
FTPSSYSSTGSNANINNANNTALGRLPRPAGYPSTVSTVEGHGPPLSPAIRVQEVAWKLS
SNRERHVPMCEDLELRRDSGSAGTQAHCLLLRKANPSRCHSRESQAAMAGQEETSQDETY
EVKMNHDTEACSEPSLLSTEMLSYQDDENRQLTLPEEDKRDIRQSPKRGFLRSASLGRRA
SFHLECLKRQKDRGGDISQKTVLPLHLVHHQALAVAGLSPLLQRSHSPASFPRPFATPPA
TPGSRGWPPQPVPTLRLEGVESSEKLNSSFPSIHCGSWAETTPGGGGSSAARRVRPVSLM
VPSQAGAPGRQFHGSASSLVEAVLISEGLGQFAQDPKFIEVTTQELADACDMTIEEMESA
ADNILSGGAPQSPNGALLPFVNCRDAGQDRAGGEEDAGCVRARGRPSEEELQDSRVYVSS
L
|
|
|
|---|
| BDBM50233393 |
|---|
| n/a |
|---|
| Name | BDBM50233393 |
|---|
| Synonyms: | CHEMBL4096195 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C29H29F3N6O3 |
|---|
| Mol. Mass. | 566.5742 |
|---|
| SMILES | CC(C)[C@]1(CC[C@](C)(C1)c1nc(-c2ccc(cc2)C(=O)Nc2cc(ccn2)C(F)(F)F)c2c(N)nccn12)C(O)=O |r| |
|---|
| Structure | 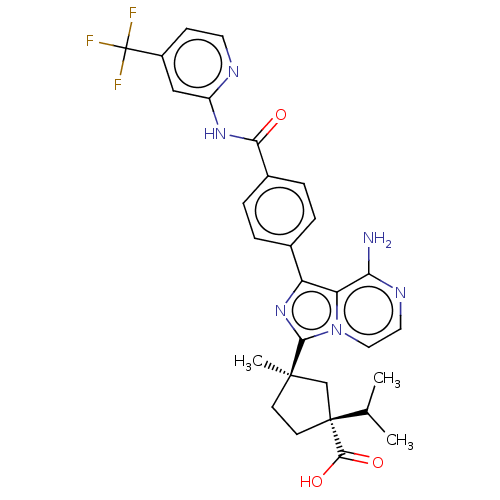
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Gao, X; Wang, J; Liu, J; Guiadeen, D; Krikorian, A; Boga, SB; Alhassan, AB; Selyutin, O; Yu, W; Yu, Y; Anand, R; Liu, S; Yang, C; Wu, H; Cai, J; Cooper, A; Zhu, H; Maloney, K; Gao, YD; Fischmann, TO; Presland, J; Mansueto, M; Xu, Z; Leccese, E; Zhang-Hoover, J; Knemeyer, I; Garlisi, CG; Bays, N; Stivers, P; Brandish, PE; Hicks, A; Kim, R; Kozlowski, JA Discovery of novel BTK inhibitors with carboxylic acids. Bioorg Med Chem Lett27:1471-1477 (2017) [PubMed] Article
Gao, X; Wang, J; Liu, J; Guiadeen, D; Krikorian, A; Boga, SB; Alhassan, AB; Selyutin, O; Yu, W; Yu, Y; Anand, R; Liu, S; Yang, C; Wu, H; Cai, J; Cooper, A; Zhu, H; Maloney, K; Gao, YD; Fischmann, TO; Presland, J; Mansueto, M; Xu, Z; Leccese, E; Zhang-Hoover, J; Knemeyer, I; Garlisi, CG; Bays, N; Stivers, P; Brandish, PE; Hicks, A; Kim, R; Kozlowski, JA Discovery of novel BTK inhibitors with carboxylic acids. Bioorg Med Chem Lett27:1471-1477 (2017) [PubMed] Article