

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | 5'-nucleotidase | ||
| Ligand | BDBM50248034 | ||
| Substrate/Competitor | n/a | ||
| Ki | 123.6±n/a nM | ||
| Comments | PDSP_1754 | ||
| Citation |  Betancur, C; Canton, M; Burgos, A; Labeeuw, B; Gully, D; Rost&egrove;ne, W; Pélaprat, D Characterization of binding sites of a new neurotensin receptor antagonist, [3H]SR 142948A, in the rat brain. Eur J Pharmacol343:67-77 (1998) [PubMed] Article Betancur, C; Canton, M; Burgos, A; Labeeuw, B; Gully, D; Rost&egrove;ne, W; Pélaprat, D Characterization of binding sites of a new neurotensin receptor antagonist, [3H]SR 142948A, in the rat brain. Eur J Pharmacol343:67-77 (1998) [PubMed] Article | ||
| More Info.: | Get all data from this article | ||
| 5'-nucleotidase | |||
| Name: | 5'-nucleotidase | ||
| Synonyms: | 5'-nucleotidase | 5NTD_RAT | Ecto-5'-nucleotidase (e5'NT) | Ecto-5-nucleotidase (e5'NT) | NT | Nt5 | Nt5e | Nte | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 63971.44 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | P21588 | ||
| Residue: | 576 | ||
| Sequence: |
| ||
| BDBM50248034 | |||
| n/a | |||
| Name | BDBM50248034 | ||
| Synonyms: | 2-{[1-(7-Chloro-quinolin-4-yl)-5-(2,6-dimethoxy-phenyl)-1H-pyrazole-3-carbonyl]-amino}-adamantane-2-carboxylic acid | CHEMBL506981 | SR 48692 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C32H31ClN4O5 | ||
| Mol. Mass. | 587.065 | ||
| SMILES | COc1cccc(OC)c1-c1cc(nn1-c1ccnc2cc(Cl)ccc12)C(=O)NC1(C2CC3CC(C2)CC1C3)C(O)=O |TLB:38:37:35:31.32.33,39:29:35:31.32.33,28:29:31.38.32:36.34.35,THB:38:32:29.37.36:35,39:29:31.38.32:36.34.35,28:29:35:31.32.33,33:32:29:36.34.35,33:34:29:31.38.32,(-7.63,.4,;-6.3,-.36,;-6.29,-1.9,;-7.62,-2.68,;-7.62,-4.22,;-6.29,-4.99,;-4.95,-4.22,;-3.61,-4.99,;-3.61,-6.53,;-4.95,-2.67,;-3.63,-1.9,;-2.22,-2.52,;-1.19,-1.37,;-1.97,-.04,;-3.47,-.36,;-4.62,.66,;-6.08,.17,;-7.22,1.19,;-6.91,2.71,;-5.45,3.19,;-5.14,4.69,;-3.68,5.16,;-3.37,6.67,;-2.53,4.14,;-2.85,2.64,;-4.31,2.16,;.35,-1.53,;.98,-2.93,;1.24,-.27,;2.78,-.43,;3.43,1.2,;4.84,1.23,;6.02,2.07,;5.46,3.5,;3.95,3.52,;2.87,2.58,;3.44,1.99,;4.02,.52,;5.54,.51,;3.42,-1.84,;2.51,-3.09,;4.95,-2,)| | ||
| Structure | 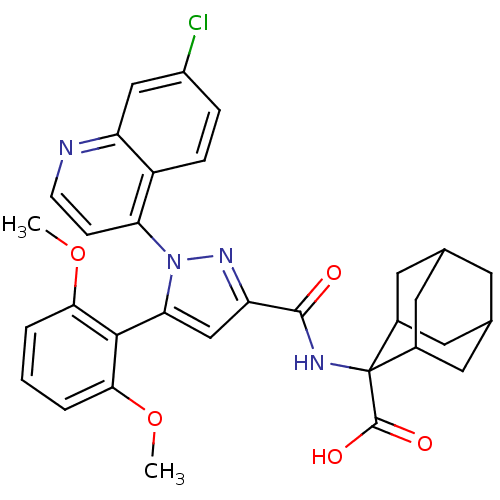
| ||