| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase |
|---|
| Ligand | BDBM85278 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Enzymatic Assay |
|---|
| pH | 8±0 |
|---|
| Temperature | 310.15±0 K |
|---|
| IC50 | 5.60e+5±n/a nM |
|---|
| Citation |  Sova, M; Kovac, A; Turk, S; Hrast, M; Blanot, D; Gobec, S Phosphorylated hydroxyethylamines as novel inhibitors of the bacterial cell wall biosynthesis enzymes MurC to MurF. Bioorg Chem37:217-22 (2009) [PubMed] Article Sova, M; Kovac, A; Turk, S; Hrast, M; Blanot, D; Gobec, S Phosphorylated hydroxyethylamines as novel inhibitors of the bacterial cell wall biosynthesis enzymes MurC to MurF. Bioorg Chem37:217-22 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase |
|---|
| Name: | UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase |
|---|
| Synonyms: | MURF_ECOLI | MurF (E. coli) | mra | murF |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 47439.03 |
|---|
| Organism: | Escherichia coli |
|---|
| Description: | P11880 |
|---|
| Residue: | 452 |
|---|
| Sequence: | MISVTLSQLTDILNGELQGADITLDAVTTDTRKLTPGCLFVALKGERFDAHDFADQAKAG
GAGALLVSRPLDIDLPQLIVKDTRLAFGELAAWVRQQVPARVVALTGSSGKTSVKEMTAA
ILSQCGNTLYTAGNLNNDIGVPMTLLRLTPEYDYAVIELGANHQGEIAWTVSLTRPEAAL
VNNLAAAHLEGFGSLAGVAKAKGEIFSGLPENGIAIMNADNNDWLNWQSVIGSRKVWRFS
PNAANSDFTATNIHVTSHGTEFTLQTPTGSVDVLLPLPGRHNIANALAAAALSMSVGATL
DAIKAGLANLKAVPGRLFPIQLAENQLLLDDSYNANVGSMTAAVQVLAEMPGYRVLVVGD
MAELGAESEACHVQVGEAAKAAGIDRVLSVGKQSHAISTASGVGEHFADKTALITRLKLL
IAEQQVITILVKGSRSAAMEEVVRALQENGTC
|
|
|
|---|
| BDBM85278 |
|---|
| n/a |
|---|
| Name | BDBM85278 |
|---|
| Synonyms: | HEA derivative, 7e |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H23N2O7PS |
|---|
| Mol. Mass. | 394.38 |
|---|
| SMILES | Cc1ccc(cc1)S(=O)(=O)NCC(CN1CCOCC1)OP(O)(O)=O |
|---|
| Structure | 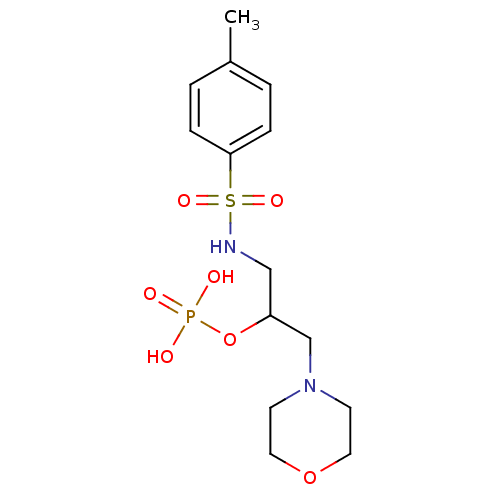
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Sova, M; Kovac, A; Turk, S; Hrast, M; Blanot, D; Gobec, S Phosphorylated hydroxyethylamines as novel inhibitors of the bacterial cell wall biosynthesis enzymes MurC to MurF. Bioorg Chem37:217-22 (2009) [PubMed] Article
Sova, M; Kovac, A; Turk, S; Hrast, M; Blanot, D; Gobec, S Phosphorylated hydroxyethylamines as novel inhibitors of the bacterial cell wall biosynthesis enzymes MurC to MurF. Bioorg Chem37:217-22 (2009) [PubMed] Article