| Reaction Details |
|---|
 | Report a problem with these data |
| Target | D(2) dopamine receptor |
|---|
| Ligand | BDBM48320 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | 64±n/a nM |
|---|
| Comments | PDSP_1233 |
|---|
| Citation |  Toll, L; Berzetei-Gurske, IP; Polgar, WE; Brandt, SR; Adapa, ID; Rodriguez, L; Schwartz, RW; Haggart, D; O'Brien, A; White, A; Kennedy, JM; Craymer, K; Farrington, L; Auh, JS Standard binding and functional assays related to medications development division testing for potential cocaine and opiate narcotic treatment medications. NIDA Res Monogr178:440-66 (1998) [PubMed] Toll, L; Berzetei-Gurske, IP; Polgar, WE; Brandt, SR; Adapa, ID; Rodriguez, L; Schwartz, RW; Haggart, D; O'Brien, A; White, A; Kennedy, JM; Craymer, K; Farrington, L; Auh, JS Standard binding and functional assays related to medications development division testing for potential cocaine and opiate narcotic treatment medications. NIDA Res Monogr178:440-66 (1998) [PubMed] |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| D(2) dopamine receptor |
|---|
| Name: | D(2) dopamine receptor |
|---|
| Synonyms: | D(2) dopamine receptor | DOPAMINE D2 | DOPAMINE D2 Long | DOPAMINE D2 Short | DRD2 | DRD2_HUMAN | Dopamine D2 receptor | Dopamine D2 receptor (D2) | Dopamine D2 receptor (D2R) | Dopamine D2A | Dopamine2-like | d2 |
|---|
| Type: | Cell-surface receptors |
|---|
| Mol. Mass.: | 50647.10 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P14416 |
|---|
| Residue: | 443 |
|---|
| Sequence: | MDPLNLSWYDDDLERQNWSRPFNGSDGKADRPHYNYYATLLTLLIAVIVFGNVLVCMAVS
REKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTA
SILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMISIVWVLSFTISCPLLFGLNNADQN
ECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRRRRKRVNTKRSSRAFRAHLRAPL
KGNCTHPEDMKLCTVIMKSNGSFPVNRRRVEAARRAQELEMEMLSSTSPPERTRYSPIPP
SHHQLTLPDPSHHGLHSTPDSPAKPEKNGHAKDHPKIAKIFEIQTMPNGKTRTSLKTMSR
RKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSA
VNPIIYTTFNIEFRKAFLKILHC
|
|
|
|---|
| BDBM48320 |
|---|
| n/a |
|---|
| Name | BDBM48320 |
|---|
| Synonyms: | 4-amino-5-chloro-N-[2-(diethylamino)ethyl]-2-methoxy-benzamide;hydrochloride | 4-amino-5-chloro-N-[2-(diethylamino)ethyl]-2-methoxybenzamide;hydrochloride | 4-azanyl-5-chloranyl-N-[2-(diethylamino)ethyl]-2-methoxy-benzamide;hydrochloride | METOCLOPRAMIDE | METOCLOPRAMIDE HYDROCHLORIDE | MLS000069667 | Metochloropramide | SMR000058471 | US11147820, Compound Metoclopramide | US11717500, Compound Metoclopramide | US9132134, Metoclopramide | cid_23659 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H22ClN3O2 |
|---|
| Mol. Mass. | 299.796 |
|---|
| SMILES | CCN(CC)CCNC(=O)c1cc(Cl)c(N)cc1OC |
|---|
| Structure | 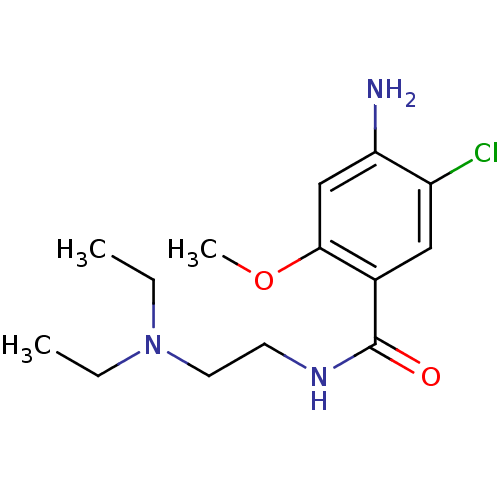
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Toll, L; Berzetei-Gurske, IP; Polgar, WE; Brandt, SR; Adapa, ID; Rodriguez, L; Schwartz, RW; Haggart, D; O'Brien, A; White, A; Kennedy, JM; Craymer, K; Farrington, L; Auh, JS Standard binding and functional assays related to medications development division testing for potential cocaine and opiate narcotic treatment medications. NIDA Res Monogr178:440-66 (1998) [PubMed]
Toll, L; Berzetei-Gurske, IP; Polgar, WE; Brandt, SR; Adapa, ID; Rodriguez, L; Schwartz, RW; Haggart, D; O'Brien, A; White, A; Kennedy, JM; Craymer, K; Farrington, L; Auh, JS Standard binding and functional assays related to medications development division testing for potential cocaine and opiate narcotic treatment medications. NIDA Res Monogr178:440-66 (1998) [PubMed]