| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prostaglandin E2 receptor EP4 subtype |
|---|
| Ligand | BDBM50085910 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | 9137±n/a nM |
|---|
| Comments | PDSP_547 |
|---|
| Citation |  Abramovitz, M; Adam, M; Boie, Y; Carrière, M; Denis, D; Godbout, C; Lamontagne, S; Rochette, C; Sawyer, N; Tremblay, NM; Belley, M; Gallant, M; Dufresne, C; Gareau, Y; Ruel, R; Juteau, H; Labelle, M; Ouimet, N; Metters, KM The utilization of recombinant prostanoid receptors to determine the affinities and selectivities of prostaglandins and related analogs. Biochim Biophys Acta1483:285-93 (2000) [PubMed] Article Abramovitz, M; Adam, M; Boie, Y; Carrière, M; Denis, D; Godbout, C; Lamontagne, S; Rochette, C; Sawyer, N; Tremblay, NM; Belley, M; Gallant, M; Dufresne, C; Gareau, Y; Ruel, R; Juteau, H; Labelle, M; Ouimet, N; Metters, KM The utilization of recombinant prostanoid receptors to determine the affinities and selectivities of prostaglandins and related analogs. Biochim Biophys Acta1483:285-93 (2000) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Prostaglandin E2 receptor EP4 subtype |
|---|
| Name: | Prostaglandin E2 receptor EP4 subtype |
|---|
| Synonyms: | PE2R4_HUMAN | PGE receptor EP4 subtype | PGE2 receptor EP4 subtype | PTGER2 | PTGER4 | Prostaglandin E2 receptor | Prostanoid EP4 receptor |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 53134.53 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P35408 |
|---|
| Residue: | 488 |
|---|
| Sequence: | MSTPGVNSSASLSPDRLNSPVTIPAVMFIFGVVGNLVAIVVLCKSRKEQKETTFYTLVCG
LAVTDLLGTLLVSPVTIATYMKGQWPGGQPLCEYSTFILLFFSLSGLSIICAMSVERYLA
INHAYFYSHYVDKRLAGLTLFAVYASNVLFCALPNMGLGSSRLQYPDTWCFIDWTTNVTA
HAAYSYMYAGFSSFLILATVLCNVLVCGALLRMHRQFMRRTSLGTEQHHAAAAASVASRG
HPAASPALPRLSDFRRRRSFRRIAGAEIQMVILLIATSLVVLICSIPLVVRVFVNQLYQP
SLEREVSKNPDLQAIRIASVNPILDPWIYILLRKTVLSKAIEKIKCLFCRIGGSRRERSG
QHCSDSQRTSSAMSGHSRSFISRELKEISSTSQTLLPDLSLPDLSENGLGGRNLLPGVPG
MGLAQEDTTSLRTLRISETSDSSQGQDSESVLLVDEAGGSGRAGPAPKGSSLQVTFPSET
LNLSEKCI
|
|
|
|---|
| BDBM50085910 |
|---|
| n/a |
|---|
| Name | BDBM50085910 |
|---|
| Synonyms: | (Z)-7-{(1R,2R,3R,5S)-2-[(E)-(R)-4-(3-Chloro-phenoxy)-3-hydroxy-but-1-enyl]-3,5-dihydroxy-cyclopentyl}-hept-5-enoic acid | 7-{2-[4-(3-Chloro-phenoxy)-3-hydroxy-but-1-enyl]-3,5-dihydroxy-cyclopentyl}-hept-5-enoic acid | CHEMBL37853 | CLOPROSTENOL |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H29ClO6 |
|---|
| Mol. Mass. | 424.915 |
|---|
| SMILES | O[C@@H](COc1cccc(Cl)c1)\C=C\[C@H]1[C@H](O)C[C@H](O)[C@@H]1C\C=C/CCCC(O)=O |
|---|
| Structure | 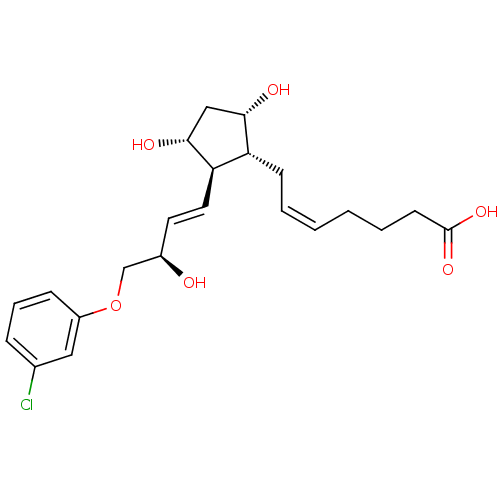
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Abramovitz, M; Adam, M; Boie, Y; Carrière, M; Denis, D; Godbout, C; Lamontagne, S; Rochette, C; Sawyer, N; Tremblay, NM; Belley, M; Gallant, M; Dufresne, C; Gareau, Y; Ruel, R; Juteau, H; Labelle, M; Ouimet, N; Metters, KM The utilization of recombinant prostanoid receptors to determine the affinities and selectivities of prostaglandins and related analogs. Biochim Biophys Acta1483:285-93 (2000) [PubMed] Article
Abramovitz, M; Adam, M; Boie, Y; Carrière, M; Denis, D; Godbout, C; Lamontagne, S; Rochette, C; Sawyer, N; Tremblay, NM; Belley, M; Gallant, M; Dufresne, C; Gareau, Y; Ruel, R; Juteau, H; Labelle, M; Ouimet, N; Metters, KM The utilization of recombinant prostanoid receptors to determine the affinities and selectivities of prostaglandins and related analogs. Biochim Biophys Acta1483:285-93 (2000) [PubMed] Article