

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Corticotropin-releasing factor receptor 1 | ||
| Ligand | BDBM50087713 | ||
| Substrate/Competitor | n/a | ||
| Ki | 1±n/a nM | ||
| Comments | PDSP_3062 | ||
| Citation |  Zhang, G; Huang, N; Li, YW; Qi, X; Marshall, AP; Yan, XX; Hill, G; Rominger, C; Prakash, SR; Bakthavatchalam, R; Rominger, DH; Gilligan, PJ; Zaczek, R Pharmacological characterization of a novel nonpeptide antagonist radioligand, (+/-)-N-[2-methyl-4-methoxyphenyl]-1-(1-(methoxymethyl) propyl)-6-methyl-1H-1,2,3-triazolo[4,5-c]pyridin-4-amine ([3H]SN003) for corticotropin-releasing factor1 receptors. J Pharmacol Exp Ther305:57-69 (2003) [PubMed] Article Zhang, G; Huang, N; Li, YW; Qi, X; Marshall, AP; Yan, XX; Hill, G; Rominger, C; Prakash, SR; Bakthavatchalam, R; Rominger, DH; Gilligan, PJ; Zaczek, R Pharmacological characterization of a novel nonpeptide antagonist radioligand, (+/-)-N-[2-methyl-4-methoxyphenyl]-1-(1-(methoxymethyl) propyl)-6-methyl-1H-1,2,3-triazolo[4,5-c]pyridin-4-amine ([3H]SN003) for corticotropin-releasing factor1 receptors. J Pharmacol Exp Ther305:57-69 (2003) [PubMed] Article | ||
| More Info.: | Get all data from this article | ||
| Corticotropin-releasing factor receptor 1 | |||
| Name: | Corticotropin-releasing factor receptor 1 | ||
| Synonyms: | CRF-R | CRF-R2 Alpha | CRF1 | CRFR | CRFR1 | CRFR1_HUMAN | CRH-R 1 | CRHR | CRHR1 | Corticotropin releasing factor receptor 1 | Corticotropin-releasing factor receptor 1 (CRF-1) | Corticotropin-releasing factor receptor 1 (CRF1) | Corticotropin-releasing hormone receptor 1 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 50744.31 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34998 | ||
| Residue: | 444 | ||
| Sequence: |
| ||
| BDBM50087713 | |||
| n/a | |||
| Name | BDBM50087713 | ||
| Synonyms: | (1-Ethyl-propyl)-[3-(4-methoxy-2-methyl-phenyl)-2,5-dimethyl-pyrazolo[1,5-a]pyrimidin-7-yl]-amine | CHEMBL45187 | DMP-904 | DMP904 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C21H28N4O | ||
| Mol. Mass. | 352.4732 | ||
| SMILES | CCC(CC)Nc1cc(C)nc2c(c(C)nn12)-c1ccc(OC)cc1C |(-4.03,1.25,;-2.68,.48,;-1.35,1.27,;-1.37,2.81,;-2.86,3.21,;-.02,.5,;-.02,-1.04,;-1.34,-1.81,;-1.34,-3.35,;-2.66,-4.12,;,-4.12,;1.33,-3.35,;2.8,-3.8,;3.7,-2.55,;5.24,-2.53,;2.77,-1.32,;1.33,-1.81,;3.3,-5.26,;2.28,-6.39,;2.77,-7.87,;4.28,-8.16,;4.78,-9.63,;3.76,-10.79,;5.3,-7,;4.79,-5.55,;5.82,-4.4,)| | ||
| Structure | 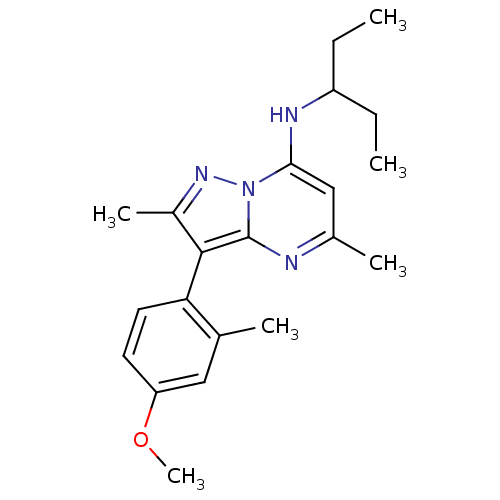
| ||