| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ATP-sensitive inward rectifier potassium channel 1 |
|---|
| Ligand | BDBM50133817 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | >10000±n/a nM |
|---|
| Comments | PDSP_7569 |
|---|
| Citation |  Valenzano, KJ; Grant, ER; Wu, G; Hachicha, M; Schmid, L; Tafesse, L; Sun, Q; Rotshteyn, Y; Francis, J; Limberis, J; Malik, S; Whittemore, ER; Hodges, D N-(4-tertiarybutylphenyl)-4-(3-chloropyridin-2-yl)tetrahydropyrazine -1(2H)-carbox-amide (BCTC), a novel, orally effective vanilloid receptor 1 antagonist with analgesic properties: I. in vitro characterization and pharmacokinetic properties. J Pharmacol Exp Ther306:377-86 (2003) [PubMed] Article Valenzano, KJ; Grant, ER; Wu, G; Hachicha, M; Schmid, L; Tafesse, L; Sun, Q; Rotshteyn, Y; Francis, J; Limberis, J; Malik, S; Whittemore, ER; Hodges, D N-(4-tertiarybutylphenyl)-4-(3-chloropyridin-2-yl)tetrahydropyrazine -1(2H)-carbox-amide (BCTC), a novel, orally effective vanilloid receptor 1 antagonist with analgesic properties: I. in vitro characterization and pharmacokinetic properties. J Pharmacol Exp Ther306:377-86 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| ATP-sensitive inward rectifier potassium channel 1 |
|---|
| Name: | ATP-sensitive inward rectifier potassium channel 1 |
|---|
| Synonyms: | ATP-regulated potassium channel ROM-K | ATP-sensitive inward rectifier potassium channel 1 | Inward rectifier K(+) channel Kir1.1 | KAB-1 | KCNJ1_RAT | Kcnj1 | Potassium channel (ATP modulatory) | Potassium channel, inwardly rectifying subfamily J member 1 | Renal Outer Medullary Potassium (ROMK) | Renal Outer Medullary Potassium (ROMK1) | Romk1 | The Renal Outer Medullary Potassium (ROMK) channel (Kir1.1) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 44976.27 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P35560 |
|---|
| Residue: | 391 |
|---|
| Sequence: | MGASERSVFRVLIRALTERMFKHLRRWFITHIFGRSRQRARLVSKEGRCNIEFGNVDAQS
RFIFFVDIWTTVLDLKWRYKMTVFITAFLGSWFLFGLLWYVVAYVHKDLPEFYPPDNRTP
CVENINGMTSAFLFSLETQVTIGYGFRFVTEQCATAIFLLIFQSILGVIINSFMCGAILA
KISRPKKRAKTITFSKNAVISKRGGKLCLLIRVANLRKSLLIGSHIYGKLLKTTITPEGE
TIILDQTNINFVVDAGNENLFFISPLTIYHIIDHNSPFFHMAAETLSQQDFELVVFLDGT
VESTSATCQVRTSYVPEEVLWGYRFVPIVSKTKEGKYRVDFHNFGKTVEVETPHCAMCLY
NEKDARARMKRGYDNPNFVLSEVDETDDTQM
|
|
|
|---|
| BDBM50133817 |
|---|
| n/a |
|---|
| Name | BDBM50133817 |
|---|
| Synonyms: | 4-(3-Chloro-pyridin-2-yl)-piperazine-1-carboxylic acid (4-tert-butyl-phenyl)-amide | BCTC | CHEMBL441472 | N-(4-tert-butylphenyl)-4-(3-chloropyridin-2-yl)piperazine-1-carboxamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H25ClN4O |
|---|
| Mol. Mass. | 372.892 |
|---|
| SMILES | CC(C)(C)c1ccc(NC(=O)N2CCN(CC2)c2ncccc2Cl)cc1 |
|---|
| Structure | 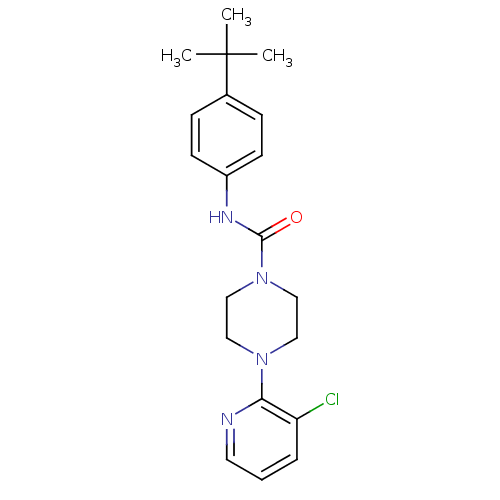
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Valenzano, KJ; Grant, ER; Wu, G; Hachicha, M; Schmid, L; Tafesse, L; Sun, Q; Rotshteyn, Y; Francis, J; Limberis, J; Malik, S; Whittemore, ER; Hodges, D N-(4-tertiarybutylphenyl)-4-(3-chloropyridin-2-yl)tetrahydropyrazine -1(2H)-carbox-amide (BCTC), a novel, orally effective vanilloid receptor 1 antagonist with analgesic properties: I. in vitro characterization and pharmacokinetic properties. J Pharmacol Exp Ther306:377-86 (2003) [PubMed] Article
Valenzano, KJ; Grant, ER; Wu, G; Hachicha, M; Schmid, L; Tafesse, L; Sun, Q; Rotshteyn, Y; Francis, J; Limberis, J; Malik, S; Whittemore, ER; Hodges, D N-(4-tertiarybutylphenyl)-4-(3-chloropyridin-2-yl)tetrahydropyrazine -1(2H)-carbox-amide (BCTC), a novel, orally effective vanilloid receptor 1 antagonist with analgesic properties: I. in vitro characterization and pharmacokinetic properties. J Pharmacol Exp Ther306:377-86 (2003) [PubMed] Article