| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Polycystin-1 |
|---|
| Ligand | BDBM86430 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | 38±n/a nM |
|---|
| Comments | PDSP_3227 |
|---|
| Citation |  Wang, QJ; Fang, TW; Yang, D; Lewin, NE; Van Lint, J; Marquez, VE; Blumberg, PM Ligand structure-activity requirements and phospholipid dependence for the binding of phorbol esters to protein kinase D. Mol Pharmacol64:1342-8 (2003) [PubMed] Article Wang, QJ; Fang, TW; Yang, D; Lewin, NE; Van Lint, J; Marquez, VE; Blumberg, PM Ligand structure-activity requirements and phospholipid dependence for the binding of phorbol esters to protein kinase D. Mol Pharmacol64:1342-8 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Polycystin-1 |
|---|
| Name: | Polycystin-1 |
|---|
| Synonyms: | PKD1 | PKD1_HUMAN | Protein Kinase D-GST |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 462537.85 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Protein Kinase D-GST 0 HUMAN::P98161 |
|---|
| Residue: | 4303 |
|---|
| Sequence: | MPPAAPARLALALGLGLWLGALAGGPGRGCGPCEPPCLCGPAPGAACRVNCSGRGLRTLG
PALRIPADATALDVSHNLLRALDVGLLANLSALAELDISNNKISTLEEGIFANLFNLSEI
NLSGNPFECDCGLAWLPRWAEEQQVRVVQPEAATCAGPGSLAGQPLLGIPLLDSGCGEEY
VACLPDNSSGTVAAVSFSAAHEGLLQPEACSAFCFSTGQGLAALSEQGWCLCGAAQPSSA
SFACLSLCSGPPPPPAPTCRGPTLLQHVFPASPGATLVGPHGPLASGQLAAFHIAAPLPV
TATRWDFGDGSAEVDAAGPAASHRYVLPGRYHVTAVLALGAGSALLGTDVQVEAAPAALE
LVCPSSVQSDESLDLSIQNRGGSGLEAAYSIVALGEEPARAVHPLCPSDTEIFPGNGHCY
RLVVEKAAWLQAQEQCQAWAGAALAMVDSPAVQRFLVSRVTRSLDVWIGFSTVQGVEVGP
APQGEAFSLESCQNWLPGEPHPATAEHCVRLGPTGWCNTDLCSAPHSYVCELQPGGPVQD
AENLLVGAPSGDLQGPLTPLAQQDGLSAPHEPVEVMVFPGLRLSREAFLTTAEFGTQELR
RPAQLRLQVYRLLSTAGTPENGSEPESRSPDNRTQLAPACMPGGRWCPGANICLPLDASC
HPQACANGCTSGPGLPGAPYALWREFLFSVPAGPPAQYSVTLHGQDVLMLPGDLVGLQHD
AGPGALLHCSPAPGHPGPRAPYLSANASSWLPHLPAQLEGTWACPACALRLLAATEQLTV
LLGLRPNPGLRLPGRYEVRAEVGNGVSRHNLSCSFDVVSPVAGLRVIYPAPRDGRLYVPT
NGSALVLQVDSGANATATARWPGGSVSARFENVCPALVATFVPGCPWETNDTLFSVVALP
WLSEGEHVVDVVVENSASRANLSLRVTAEEPICGLRATPSPEARVLQGVLVRYSPVVEAG
SDMVFRWTINDKQSLTFQNVVFNVIYQSAAVFKLSLTASNHVSNVTVNYNVTVERMNRMQ
GLQVSTVPAVLSPNATLALTAGVLVDSAVEVAFLWTFGDGEQALHQFQPPYNESFPVPDP
SVAQVLVEHNVMHTYAAPGEYLLTVLASNAFENLTQQVPVSVRASLPSVAVGVSDGVLVA
GRPVTFYPHPLPSPGGVLYTWDFGDGSPVLTQSQPAANHTYASRGTYHVRLEVNNTVSGA
AAQADVRVFEELRGLSVDMSLAVEQGAPVVVSAAVQTGDNITWTFDMGDGTVLSGPEATV
EHVYLRAQNCTVTVGAASPAGHLARSLHVLVFVLEVLRVEPAACIPTQPDARLTAYVTGN
PAHYLFDWTFGDGSSNTTVRGCPTVTHNFTRSGTFPLALVLSSRVNRAHYFTSICVEPEV
GNVTLQPERQFVQLGDEAWLVACAWPPFPYRYTWDFGTEEAAPTRARGPEVTFIYRDPGS
YLVTVTASNNISAANDSALVEVQEPVLVTSIKVNGSLGLELQQPYLFSAVGRGRPASYLW
DLGDGGWLEGPEVTHAYNSTGDFTVRVAGWNEVSRSEAWLNVTVKRRVRGLVVNASRTVV
PLNGSVSFSTSLEAGSDVRYSWVLCDRCTPIPGGPTISYTFRSVGTFNIIVTAENEVGSA
QDSIFVYVLQLIEGLQVVGGGRYFPTNHTVQLQAVVRDGTNVSYSWTAWRDRGPALAGSG
KGFSLTVLEAGTYHVQLRATNMLGSAWADCTMDFVEPVGWLMVAASPNPAAVNTSVTLSA
ELAGGSGVVYTWSLEEGLSWETSEPFTTHSFPTPGLHLVTMTAGNPLGSANATVEVDVQV
PVSGLSIRASEPGGSFVAAGSSVPFWGQLATGTNVSWCWAVPGGSSKRGPHVTMVFPDAG
TFSIRLNASNAVSWVSATYNLTAEEPIVGLVLWASSKVVAPGQLVHFQILLAAGSAVTFR
LQVGGANPEVLPGPRFSHSFPRVGDHVVSVRGKNHVSWAQAQVRIVVLEAVSGLQVPNCC
EPGIATGTERNFTARVQRGSRVAYAWYFSLQKVQGDSLVILSGRDVTYTPVAAGLLEIQV
RAFNALGSENRTLVLEVQDAVQYVALQSGPCFTNRSAQFEAATSPSPRRVAYHWDFGDGS
PGQDTDEPRAEHSYLRPGDYRVQVNASNLVSFFVAQATVTVQVLACREPEVDVVLPLQVL
MRRSQRNYLEAHVDLRDCVTYQTEYRWEVYRTASCQRPGRPARVALPGVDVSRPRLVLPR
LALPVGHYCFVFVVSFGDTPLTQSIQANVTVAPERLVPIIEGGSYRVWSDTRDLVLDGSE
SYDPNLEDGDQTPLSFHWACVASTQREAGGCALNFGPRGSSTVTIPRERLAAGVEYTFSL
TVWKAGRKEEATNQTVLIRSGRVPIVSLECVSCKAQAVYEVSRSSYVYLEGRCLNCSSGS
KRGRWAARTFSNKTLVLDETTTSTGSAGMRLVLRRGVLRDGEGYTFTLTVLGRSGEEEGC
ASIRLSPNRPPLGGSCRLFPLGAVHALTTKVHFECTGWHDAEDAGAPLVYALLLRRCRQG
HCEEFCVYKGSLSSYGAVLPPGFRPHFEVGLAVVVQDQLGAAVVALNRSLAITLPEPNGS
ATGLTVWLHGLTASVLPGLLRQADPQHVIEYSLALVTVLNEYERALDVAAEPKHERQHRA
QIRKNITETLVSLRVHTVDDIQQIAAALAQCMGPSRELVCRSCLKQTLHKLEAMMLILQA
ETTAGTVTPTAIGDSILNITGDLIHLASSDVRAPQPSELGAESPSRMVASQAYNLTSALM
RILMRSRVLNEEPLTLAGEEIVAQGKRSDPRSLLCYGGAPGPGCHFSIPEAFSGALANLS
DVVQLIFLVDSNPFPFGYISNYTVSTKVASMAFQTQAGAQIPIERLASERAITVKVPNNS
DWAARGHRSSANSANSVVVQPQASVGAVVTLDSSNPAAGLHLQLNYTLLDGHYLSEEPEP
YLAVYLHSEPRPNEHNCSASRRIRPESLQGADHRPYTFFISPGSRDPAGSYHLNLSSHFR
WSALQVSVGLYTSLCQYFSEEDMVWRTEGLLPLEETSPRQAVCLTRHLTAFGASLFVPPS
HVRFVFPEPTADVNYIVMLTCAVCLVTYMVMAAILHKLDQLDASRGRAIPFCGQRGRFKY
EILVKTGWGRGSGTTAHVGIMLYGVDSRSGHRHLDGDRAFHRNSLDIFRIATPHSLGSVW
KIRVWHDNKGLSPAWFLQHVIVRDLQTARSAFFLVNDWLSVETEANGGLVEKEVLAASDA
ALLRFRRLLVAELQRGFFDKHIWLSIWDRPPRSRFTRIQRATCCVLLICLFLGANAVWYG
AVGDSAYSTGHVSRLSPLSVDTVAVGLVSSVVVYPVYLAILFLFRMSRSKVAGSPSPTPA
GQQVLDIDSCLDSSVLDSSFLTFSGLHAEQAFVGQMKSDLFLDDSKSLVCWPSGEGTLSW
PDLLSDPSIVGSNLRQLARGQAGHGLGPEEDGFSLASPYSPAKSFSASDEDLIQQVLAEG
VSSPAPTQDTHMETDLLSSLSSTPGEKTETLALQRLGELGPPSPGLNWEQPQAARLSRTG
LVEGLRKRLLPAWCASLAHGLSLLLVAVAVAVSGWVGASFPPGVSVAWLLSSSASFLASF
LGWEPLKVLLEALYFSLVAKRLHPDEDDTLVESPAVTPVSARVPRVRPPHGFALFLAKEE
ARKVKRLHGMLRSLLVYMLFLLVTLLASYGDASCHGHAYRLQSAIKQELHSRAFLAITRS
EELWPWMAHVLLPYVHGNQSSPELGPPRLRQVRLQEALYPDPPGPRVHTCSAAGGFSTSD
YDVGWESPHNGSGTWAYSAPDLLGAWSWGSCAVYDSGGYVQELGLSLEESRDRLRFLQLH
NWLDNRSRAVFLELTRYSPAVGLHAAVTLRLEFPAAGRALAALSVRPFALRRLSAGLSLP
LLTSVCLLLFAVHFAVAEARTWHREGRWRVLRLGAWARWLLVALTAATALVRLAQLGAAD
RQWTRFVRGRPRRFTSFDQVAQLSSAARGLAASLLFLLLVKAAQQLRFVRQWSVFGKTLC
RALPELLGVTLGLVVLGVAYAQLAILLVSSCVDSLWSVAQALLVLCPGTGLSTLCPAESW
HLSPLLCVGLWALRLWGALRLGAVILRWRYHALRGELYRPAWEPQDYEMVELFLRRLRLW
MGLSKVKEFRHKVRFEGMEPLPSRSSRGSKVSPDVPPPSAGSDASHPSTSSSQLDGLSVS
LGRLGTRCEPEPSRLQAVFEALLTQFDRLNQATEDVYQLEQQLHSLQGRRSSRAPAGSSR
GPSPGLRPALPSRLARASRGVDLATGPSRTPLRAKNKVHPSST
|
|
|
|---|
| BDBM86430 |
|---|
| n/a |
|---|
| Name | BDBM86430 |
|---|
| Synonyms: | (-)-Indolactam V | (-)-Indolactam V (low PS) | CAS_105000 | NSC_105000 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H23N3O2 |
|---|
| Mol. Mass. | 301.3834 |
|---|
| SMILES | CC(C)C1N(C)c2cccc3[nH]cc(CC(CO)NC1=O)c23 |
|---|
| Structure | 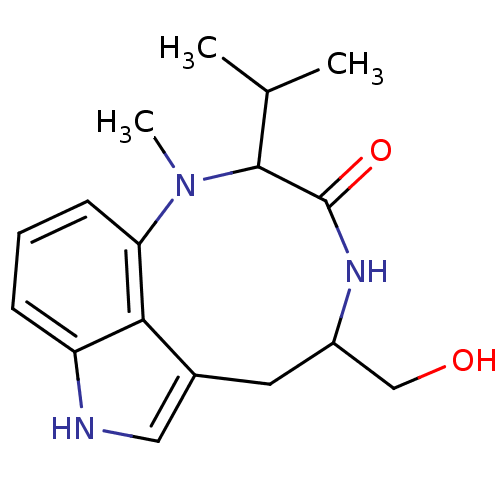
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wang, QJ; Fang, TW; Yang, D; Lewin, NE; Van Lint, J; Marquez, VE; Blumberg, PM Ligand structure-activity requirements and phospholipid dependence for the binding of phorbol esters to protein kinase D. Mol Pharmacol64:1342-8 (2003) [PubMed] Article
Wang, QJ; Fang, TW; Yang, D; Lewin, NE; Van Lint, J; Marquez, VE; Blumberg, PM Ligand structure-activity requirements and phospholipid dependence for the binding of phorbol esters to protein kinase D. Mol Pharmacol64:1342-8 (2003) [PubMed] Article