

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Isocitrate dehydrogenase [NADP] cytoplasmic [R132C] | ||
| Ligand | BDBM278540 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Inhibitory Activity Against IDH1R132H and IDH1R132C Enzymes | ||
| pH | 7.5±n/a | ||
| Temperature | 298.15±n/a K | ||
| IC50 | 47±n/a nM | ||
| Comments | extracted | ||
| Citation |  Saito, S; Itoh, M; Fujisawa, T; Saito, H; Kiyotsuka, Y; Watanabe, H; Matsunaga, H; Kagoshima, Y; Suzuki, T; Ogawara, Y; Kitabayashi, K Isoxazole derivative as mutant isocitrate dehydrogenase 1 inhibitor US Patent US10040791 Publication Date 8/7/2018 Saito, S; Itoh, M; Fujisawa, T; Saito, H; Kiyotsuka, Y; Watanabe, H; Matsunaga, H; Kagoshima, Y; Suzuki, T; Ogawara, Y; Kitabayashi, K Isoxazole derivative as mutant isocitrate dehydrogenase 1 inhibitor US Patent US10040791 Publication Date 8/7/2018 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Isocitrate dehydrogenase [NADP] cytoplasmic [R132C] | |||
| Name: | Isocitrate dehydrogenase [NADP] cytoplasmic [R132C] | ||
| Synonyms: | IDH1 | IDH1 R132C | IDH1(R132C) | IDHC_HUMAN | Isocitrate dehydrogenase (IDH1)(R132C) | Isocitrate dehydrogenase 1 mutant (R132C) | Isocitrate dehydrogenase [NADP] cytoplasmic (IDH1) (R132C) | PICD | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 46607.23 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | O75874[R132C] | ||
| Residue: | 414 | ||
| Sequence: |
| ||
| BDBM278540 | |||
| n/a | |||
| Name | BDBM278540 | ||
| Synonyms: | (2E)-3-(3-Methyl-1-{[5-(3-methyloxetan-3-yl)-3-(2,4,6-trichlorophenyl)-1,2-oxazol-4-yl]carbonyl}-1H-indol-4-yl)prop-2-enoic acid | US10040791, Example 2 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H19Cl3N2O5 | ||
| Mol. Mass. | 545.798 | ||
| SMILES | Cc1cn(C(=O)c2c(noc2C2(C)COC2)-c2c(Cl)cc(Cl)cc2Cl)c2cccc(\C=C\C(O)=O)c12 |(3.32,.91,;1.98,.14,;.52,.62,;-.39,-.63,;-1.93,-.63,;-2.57,-1.74,;-2.7,.7,;-4.24,.7,;-4.71,2.17,;-3.47,3.07,;-2.22,2.17,;-.89,2.94,;.45,2.17,;.2,4.03,;-.89,5.12,;-1.98,4.03,;-5.28,-1.12,;-6.82,-1.12,;-7.59,.22,;-7.59,-2.45,;-6.82,-3.78,;-7.59,-5.12,;-5.28,-3.78,;-4.51,-2.45,;-2.97,-2.45,;.52,-1.87,;.2,-3.38,;1.34,-4.41,;2.81,-3.94,;3.13,-2.43,;4.61,-2.03,;5.01,-.54,;6.5,-.14,;7.59,-1.23,;6.9,1.34,;1.98,-1.4,)| | ||
| Structure | 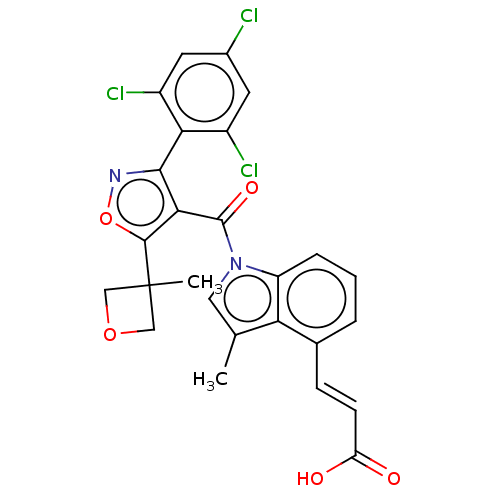
| ||