| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phosphodiesterase |
|---|
| Ligand | BDBM14361 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Enzymatic Assay |
|---|
| pH | 7.5±0 |
|---|
| Temperature | 297.15±0 K |
|---|
| IC50 | >1.00e+5±n/a nM |
|---|
| Citation |  Wang, H; Kunz, S; Chen, G; Seebeck, T; Wan, Y; Robinson, H; Martinelli, S; Ke, H Biological and structural characterization of Trypanosoma cruzi phosphodiesterase C and Implications for design of parasite selective inhibitors. J Biol Chem287:11788-97 (2012) [PubMed] Article Wang, H; Kunz, S; Chen, G; Seebeck, T; Wan, Y; Robinson, H; Martinelli, S; Ke, H Biological and structural characterization of Trypanosoma cruzi phosphodiesterase C and Implications for design of parasite selective inhibitors. J Biol Chem287:11788-97 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Phosphodiesterase |
|---|
| Name: | Phosphodiesterase |
|---|
| Synonyms: | Cyclic nucleotide specific phosphodiesterase | Phosphodiesterase (TcrPDEC) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 103074.59 |
|---|
| Organism: | Trypanosoma cruzi |
|---|
| Description: | Q53I59 |
|---|
| Residue: | 924 |
|---|
| Sequence: | MSEDAGLPVPRSQWVERGVSCATCGKRFSLFTAKSNCPCCGKLCCSDCVQAECAIVGGSA
PSKVCIDCFSMLQSRRRVEPDEGSSFREFNAASAFPLQTRLLADGRVESGETSRVSPPND
GRVQHVSRANGYSNSLPVLDEYVDDLLRKSELLRMENDVLLNRLREQEAEIHALRLERDR
AVARIVPDGGSMAGRSGLPQVSDEIVKELRGELAVAHLRIESVKRELKNALDRAKSSETM
VRNLKQGLCNYKEEVVRPLQSREEVEMLPGVNGRRDMISTRRLPPSIVQDTILAVVPPKS
CAAIGTDVDLRDWGFDTFEVASRVPSVLQSVAMHVALAWNFFASQEEAQKWAFLVAAVEN
NYRPNPYHNAIHAADVLQGTFSLVSAAKPLMEHLTPLECKAAAFAALTHDVCHPGRTNAF
LAAVQDPVSFKFSGKGTLEQLHTVTAFELLNVTEFDFTSSMDNASFLEFKNIVSHLIGHT
DMSLHSETIAKHGAKLSAGGFDCTCKEDRLEALSLLLHAADIGASSRGVAIARKWLVILQ
EFADQAEDERRRGLPVTPGFDTPSSVEKSQIPFLDFFVIPTFDLLHQLFPSIEEPLHNLR
KLRELYAAKAGVTTPFPPPVDYRSREERIRSLEAELAYFRRREEEFHRQLQELRTASENE
NKSSAAPMTREGALNKQSQLVCRGEGNMNADWADAGGGFRNGKDVRDLQDVSMDHLVSSK
TVESDTGDSGPRGRRGSKAETTKAYERKLGEREAALMATARLLENREKRLAVFSEKLAEI
AEGLHEERKRLQPMEEFKTPCFSRETELISEESASMDVTHRFSTQWEAEERLARKYRELD
ELLLIVRAMRMGYAARRNTNLGWKALSATLAEREAAISEAVELSRQRRRHLEEGRGPHPT
ATHLDRLENATFQLMSAITLLTQC
|
|
|
|---|
| BDBM14361 |
|---|
| n/a |
|---|
| Name | BDBM14361 |
|---|
| Synonyms: | (R,S)-Rolipram | 4-(3-cyclopentyloxy-4-methoxy-phenyl)pyrrolidin-2-one | 4-[3-(cyclopentyloxy)-4-methoxyphenyl]pyrrolidin-2-one | Adeo | CHEMBL63 | Rolipram |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H21NO3 |
|---|
| Mol. Mass. | 275.3428 |
|---|
| SMILES | COc1ccc(cc1OC1CCCC1)C1CNC(=O)C1 |
|---|
| Structure | 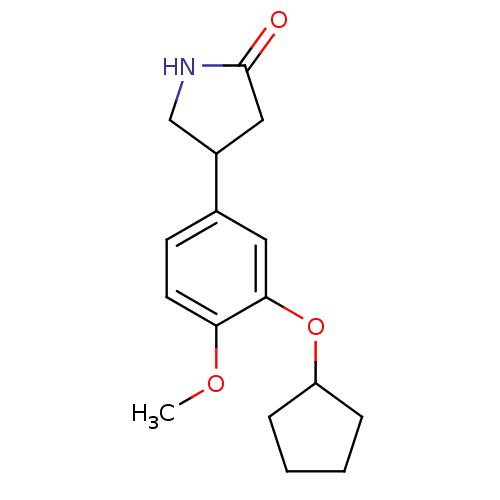
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wang, H; Kunz, S; Chen, G; Seebeck, T; Wan, Y; Robinson, H; Martinelli, S; Ke, H Biological and structural characterization of Trypanosoma cruzi phosphodiesterase C and Implications for design of parasite selective inhibitors. J Biol Chem287:11788-97 (2012) [PubMed] Article
Wang, H; Kunz, S; Chen, G; Seebeck, T; Wan, Y; Robinson, H; Martinelli, S; Ke, H Biological and structural characterization of Trypanosoma cruzi phosphodiesterase C and Implications for design of parasite selective inhibitors. J Biol Chem287:11788-97 (2012) [PubMed] Article