| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Oxaloacetate decarboxylase |
|---|
| Ligand | BDBM92966 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Inhibition Assay |
|---|
| Ki | 560000±40000 nM |
|---|
| Citation |  Narayanan, BC; Niu, W; Han, Y; Zou, J; Mariano, PS; Dunaway-Mariano, D; Herzberg, O Structure and function of PA4872 from Pseudomonas aeruginosa, a novel class of oxaloacetate decarboxylase from the PEP mutase/isocitrate lyase superfamily. Biochemistry47:167-82 (2008) [PubMed] Article Narayanan, BC; Niu, W; Han, Y; Zou, J; Mariano, PS; Dunaway-Mariano, D; Herzberg, O Structure and function of PA4872 from Pseudomonas aeruginosa, a novel class of oxaloacetate decarboxylase from the PEP mutase/isocitrate lyase superfamily. Biochemistry47:167-82 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Oxaloacetate decarboxylase |
|---|
| Name: | Oxaloacetate decarboxylase |
|---|
| Synonyms: | OADC_PSEAE | Oxaloacetate decarboxylase (PA4872) | Oxaloacetate decarboxylase (PA4872) Y212F |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 31314.95 |
|---|
| Organism: | Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 / 1C / PRS 101 / LMG 12228) |
|---|
| Description: | Q9HUU1 |
|---|
| Residue: | 287 |
|---|
| Sequence: | MHRASHHELRAMFRALLDSSRCYHTASVFDPMSARIAADLGFECGILGGSVASLQVLAAP
DFALITLSEFVEQATRIGRVARLPVIADADHGYGNALNVMRTVVELERAGIAALTIEDTL
LPAQFGRKSTDLICVEEGVGKIRAALEARVDPALTIIARTNAELIDVDAVIQRTLAYQEA
GADGICLVGVRDFAHLEAIAEHLHIPLMLVTYGNPQLRDDARLARLGVRVVVNGHAAYFA
AIKATYDCLREERGAVASDLTASELSKKYTFPEEYQAWARDYMEVKE
|
|
|
|---|
| BDBM92966 |
|---|
| n/a |
|---|
| Name | BDBM92966 |
|---|
| Synonyms: | Acetopyruvate |
|---|
| Type | Small molecule |
|---|
| Emp. Form. | C5H5O4 |
|---|
| Mol. Mass. | 129.0913 |
|---|
| SMILES | CC(=O)CC(=O)C([O-])=O |
|---|
| Structure | 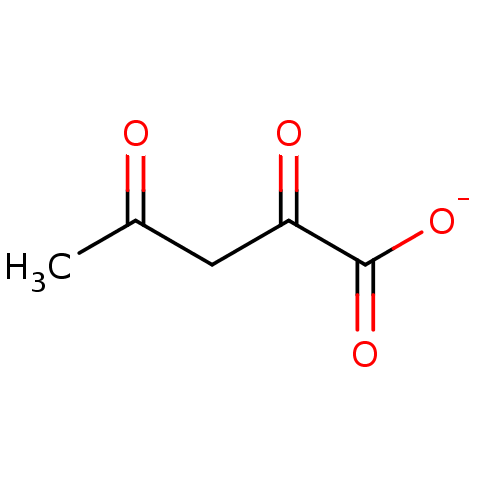
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Narayanan, BC; Niu, W; Han, Y; Zou, J; Mariano, PS; Dunaway-Mariano, D; Herzberg, O Structure and function of PA4872 from Pseudomonas aeruginosa, a novel class of oxaloacetate decarboxylase from the PEP mutase/isocitrate lyase superfamily. Biochemistry47:167-82 (2008) [PubMed] Article
Narayanan, BC; Niu, W; Han, Y; Zou, J; Mariano, PS; Dunaway-Mariano, D; Herzberg, O Structure and function of PA4872 from Pseudomonas aeruginosa, a novel class of oxaloacetate decarboxylase from the PEP mutase/isocitrate lyase superfamily. Biochemistry47:167-82 (2008) [PubMed] Article