| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prolyl hydroxylase EGLN3 |
|---|
| Ligand | BDBM93422 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | PHD Enzymatic Assay |
|---|
| IC50 | 4.5±0.0 nM |
|---|
| Citation |  Murray, JK; Balan, C; Allgeier, AM; Kasparian, A; Viswanadhan, V; Wilde, C; Allen, JR; Yoder, SC; Biddlecome, G; Hungate, RW; Miranda, LP Dipeptidyl-quinolone derivatives inhibit hypoxia inducible factor-1a prolyl hydroxylases-1, -2, and -3 with altered selectivity. J Comb Chem12:676-86 (2010) [PubMed] Article Murray, JK; Balan, C; Allgeier, AM; Kasparian, A; Viswanadhan, V; Wilde, C; Allen, JR; Yoder, SC; Biddlecome, G; Hungate, RW; Miranda, LP Dipeptidyl-quinolone derivatives inhibit hypoxia inducible factor-1a prolyl hydroxylases-1, -2, and -3 with altered selectivity. J Comb Chem12:676-86 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prolyl hydroxylase EGLN3 |
|---|
| Name: | Prolyl hydroxylase EGLN3 |
|---|
| Synonyms: | EGLN3 | EGLN3_HUMAN | Egl nine homolog 3 (EGLIN3) | Egl nine homolog 3 (EGLN3) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 27265.54 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9H6Z9 |
|---|
| Residue: | 239 |
|---|
| Sequence: | MPLGHIMRLDLEKIALEYIVPCLHEVGFCYLDNFLGEVVGDCVLERVKQLHCTGALRDGQ
LAGPRAGVSKRHLRGDQITWIGGNEEGCEAISFLLSLIDRLVLYCGSRLGKYYVKERSKA
MVACYPGNGTGYVRHVDNPNGDGRCITCIYYLNKNWDAKLHGGILRIFPEGKSFIADVEP
IFDRLLFFWSDRRNPHEVQPSYATRYAMTVWYFDAEERAEAKKKFRNLTRKTESALTED
|
|
|
|---|
| BDBM93422 |
|---|
| n/a |
|---|
| Name | BDBM93422 |
|---|
| Synonyms: | PHD Inhibitor, 12{1,1,2} |
|---|
| Type | Small molecule |
|---|
| Emp. Form. | C38H37N5O8 |
|---|
| Mol. Mass. | 691.7291 |
|---|
| SMILES | CC(=O)N[C@@H](Cc1ccc2ccccc2c1)C(=O)N[C@@H](Cc1ccccc1)C(=O)NCc1ccc2n(C)c(=O)c(C(=O)NCC(O)=O)c(O)c2c1 |r| |
|---|
| Structure | 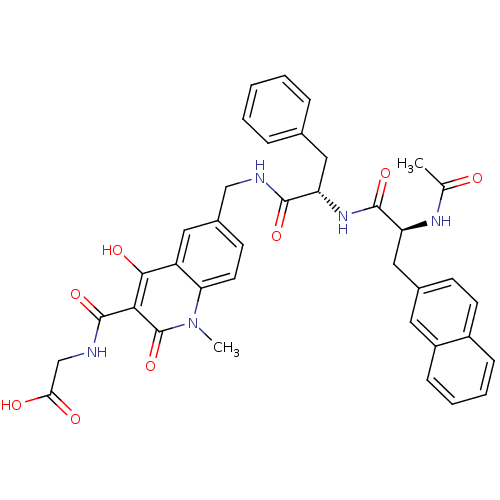
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Murray, JK; Balan, C; Allgeier, AM; Kasparian, A; Viswanadhan, V; Wilde, C; Allen, JR; Yoder, SC; Biddlecome, G; Hungate, RW; Miranda, LP Dipeptidyl-quinolone derivatives inhibit hypoxia inducible factor-1a prolyl hydroxylases-1, -2, and -3 with altered selectivity. J Comb Chem12:676-86 (2010) [PubMed] Article
Murray, JK; Balan, C; Allgeier, AM; Kasparian, A; Viswanadhan, V; Wilde, C; Allen, JR; Yoder, SC; Biddlecome, G; Hungate, RW; Miranda, LP Dipeptidyl-quinolone derivatives inhibit hypoxia inducible factor-1a prolyl hydroxylases-1, -2, and -3 with altered selectivity. J Comb Chem12:676-86 (2010) [PubMed] Article