| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Nuclear receptor subfamily 5 group A member 2 |
|---|
| Ligand | BDBM52729 |
|---|
| Substrate/Competitor | n/a |
|---|
| IC50 | 8942±n/a nM |
|---|
| Citation |  PubChem, PC Luminescence-based cell-based high throughput dose response assay for inverse agonists of the liver receptor homolog-1 (LRH-1; NR5A2) PubChem Bioassay(2013)[AID] PubChem, PC Luminescence-based cell-based high throughput dose response assay for inverse agonists of the liver receptor homolog-1 (LRH-1; NR5A2) PubChem Bioassay(2013)[AID] |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Nuclear receptor subfamily 5 group A member 2 |
|---|
| Name: | Nuclear receptor subfamily 5 group A member 2 |
|---|
| Synonyms: | Alpha-1-fetoprotein transcription factor | B1-binding factor | B1F | CPF | CYP7A promoter-binding factor | FTF | Hepatocytic transcription factor | LRH-1 | Liver Receptor Homolog 1 | NR5A2 | NR5A2_HUMAN | Nuclear receptor subfamily 5 group A member 2 | hB1F | nuclear receptor subfamily 5 group A member 2 isoform 2 |
|---|
| Type: | Nuclear Hormone Receptor |
|---|
| Mol. Mass.: | 61341.37 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 541 |
|---|
| Sequence: | MSSNSDTGDLQESLKHGLTPIGAGLPDRHGSPIPARGRLVMLPKVETEALGLARSHGEQG
QMPENMQVSQFKMVNYSYDEDLEELCPVCGDKVSGYHYGLLTCESCKGFFKRTVQNNKRY
TCIENQNCQIDKTQRKRCPYCRFQKCLSVGMKLEAVRADRMRGGRNKFGPMYKRDRALKQ
QKKALIRANGLKLEAMSQVIQAMPSDLTISSAIQNIHSASKGLPLNHAALPPTDYDRSPF
VTSPISMTMPPHGSLQGYQTYGHFPSRAIKSEYPDPYTSSPESIMGYSYMDSYQTSSPAS
IPHLILELLKCEPDEPQVQAKIMAYLQQEQANRSKHEKLSTFGLMCKMADQTLFSIVEWA
RSSIFFRELKVDDQMKLLQNCWSELLILDHIYRQVVHGKEGSIFLVTGQQVDYSIIASQA
GATLNNLMSHAQELVAKLRSLQFDQREFVCLKFLVLFSLDVKNLENFQLVEGVQEQVNAA
LLDYTMCNYPQQTEKFGQLLLRLPEIRAISMQAEEYLYYKHLNGDVPYNNLLIEMLHAKR
A
|
|
|
|---|
| BDBM52729 |
|---|
| n/a |
|---|
| Name | BDBM52729 |
|---|
| Synonyms: | 2-chloro-N-[2-[(5Z)-2,4-diketo-5-(3-pyridylmethylene)thiazolidin-3-yl]ethyl]acetamide | 2-chloro-N-[2-[(5Z)-2,4-dioxo-5-(3-pyridinylmethylidene)-3-thiazolidinyl]ethyl]acetamide | 2-chloro-N-[2-[(5Z)-2,4-dioxo-5-(pyridin-3-ylmethylidene)-1,3-thiazolidin-3-yl]ethyl]acetamide | MLS000517206 | N-[2-[(5Z)-2,4-bis(oxidanylidene)-5-(pyridin-3-ylmethylidene)-1,3-thiazolidin-3-yl]ethyl]-2-chloranyl-ethanamide | SMR000343357 | cid_2434777 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H12ClN3O3S |
|---|
| Mol. Mass. | 325.771 |
|---|
| SMILES | ClCC(=O)NCCN1C(=O)S\C(=C/c2cccnc2)C1=O |
|---|
| Structure | 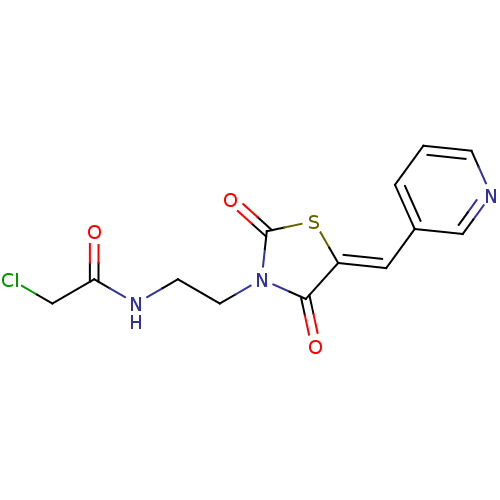
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Luminescence-based cell-based high throughput dose response assay for inverse agonists of the liver receptor homolog-1 (LRH-1; NR5A2) PubChem Bioassay(2013)[AID]
PubChem, PC Luminescence-based cell-based high throughput dose response assay for inverse agonists of the liver receptor homolog-1 (LRH-1; NR5A2) PubChem Bioassay(2013)[AID]