| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Kallikrein-7 |
|---|
| Ligand | BDBM51528 |
|---|
| Substrate/Competitor | n/a |
|---|
| EC50 | 3623±n/a nM |
|---|
| Citation |  PubChem, PC Fluorescence Intensity-based biochemical primary high throughput dose response assay to identify activators of kallikrein-7 (K7) zymogen PubChem Bioassay(2013)[AID] PubChem, PC Fluorescence Intensity-based biochemical primary high throughput dose response assay to identify activators of kallikrein-7 (K7) zymogen PubChem Bioassay(2013)[AID] |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Kallikrein-7 |
|---|
| Name: | Kallikrein-7 |
|---|
| Synonyms: | KLK7 | KLK7_HUMAN | Kallikrein 7 | Kallikrein-7 | PRSS6 | SCCE | Serine protease 6 | Stratum corneum chymotryptic enzyme | hK7 | hSCCE | kallikrein-7 isoform 1 preproprotein |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 27535.05 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1469257 |
|---|
| Residue: | 253 |
|---|
| Sequence: | MARSLLLPLQILLLSLALETAGEEAQGDKIIDGAPCARGSHPWQVALLSGNQLHCGGVLV
NERWVLTAAHCKMNEYTVHLGSDTLGDRRAQRIKASKSFRHPGYSTQTHVNDLMLVKLNS
QARLSSMVKKVRLPSRCEPPGTTCTVSGWGTTTSPDVTFPSDLMCVDVKLISPQDCTKVY
KDLLENSMLCAGIPDSKKNACNGDSGGPLVCRGTLQGLVSWGTFPCGQPNDPGVYTQVCK
FTKWINDTMKKHR
|
|
|
|---|
| BDBM51528 |
|---|
| n/a |
|---|
| Name | BDBM51528 |
|---|
| Synonyms: | 2-amino-1-(2-piperidinoethyl)pyrrolo[3,2-b]quinoxaline-3-carboxylic acid ethyl ester | 2-amino-1-[2-(1-piperidinyl)ethyl]-3-pyrrolo[3,2-b]quinoxalinecarboxylic acid ethyl ester | MLS000056772 | SMR000065736 | cid_1888166 | ethyl 2-amino-1-(2-piperidin-1-ylethyl)pyrrolo[3,2-b]quinoxaline-3-carboxylate | ethyl 2-azanyl-1-(2-piperidin-1-ylethyl)pyrrolo[3,2-b]quinoxaline-3-carboxylate |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H25N5O2 |
|---|
| Mol. Mass. | 367.4448 |
|---|
| SMILES | CCOC(=O)c1c(N)n(CCN2CCCCC2)c2nc3ccccc3nc12 |
|---|
| Structure | 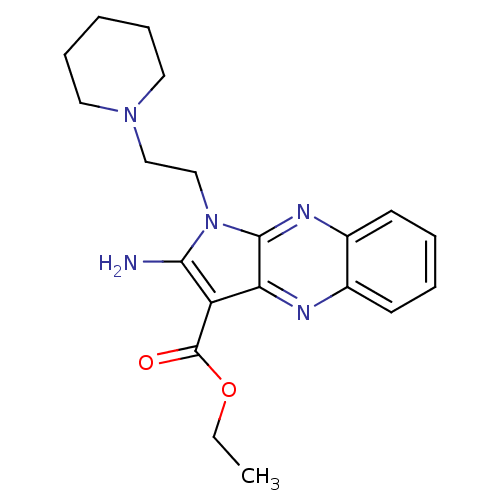
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Fluorescence Intensity-based biochemical primary high throughput dose response assay to identify activators of kallikrein-7 (K7) zymogen PubChem Bioassay(2013)[AID]
PubChem, PC Fluorescence Intensity-based biochemical primary high throughput dose response assay to identify activators of kallikrein-7 (K7) zymogen PubChem Bioassay(2013)[AID]