| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Urease [D459Y,K653P] |
|---|
| Ligand | BDBM50099857 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Urease Inhibition Assay |
|---|
| pH | 8.2±n/a |
|---|
| Temperature | 310.15±n/a K |
|---|
| IC50 | 4.32e+4± 2.6e+3 nM |
|---|
| Comments | extracted |
|---|
| Citation |  Saeed, A; Khan, MS; Rafique, H; Shahid, M; Iqbal, J Design, synthesis, molecular docking studies and in vitro screening of ethyl 4-(3-benzoylthioureido) benzoates as urease inhibitors. Bioorg Chem52:1-7 (2014) [PubMed] Article Saeed, A; Khan, MS; Rafique, H; Shahid, M; Iqbal, J Design, synthesis, molecular docking studies and in vitro screening of ethyl 4-(3-benzoylthioureido) benzoates as urease inhibitors. Bioorg Chem52:1-7 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Urease [D459Y,K653P] |
|---|
| Name: | Urease [D459Y,K653P] |
|---|
| Synonyms: | UREA_CANEN | Urea amidohydrolase | Urease |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 90763.23 |
|---|
| Organism: | Canavalia ensiformis (Jack bean) |
|---|
| Description: | P07374[D459Y,K653P] |
|---|
| Residue: | 840 |
|---|
| Sequence: | MKLSPREVEKLGLHNAGYLAQKRLARGVRLNYTEAVALIASQIMEYARDGEKTVAQLMCL
GQHLLGRRQVLPAVPHLLNAVQVEATFPDGTKLVTVHDPISRENGELQEALFGSLLPVPS
LDKFAETKEDNRIPGEILCEDECLTLNIGRKAVILKVTSKGDRPIQVGSHYHFIEVNPYL
TFDRRKAYGMRLNIAAGTAVRFEPGDCKSVTLVSIEGNKVIRGGNAIADGPVNETNLEAA
MHAVRSKGFGHEEEKDASEGFTKEDPNCPFNTFIHRKEYANKYGPTTGDKIRLGDTNLLA
EIEKDYALYGDECVFGGGKVIRDGMGQSCGHPPAISLDTVITNAVIIDYTGIIKADIGIK
DGLIASIGKAGNPDIMNGVFSNMIIGANTEVIAGEGLIVTAGAIDCHVHYICPQLVYEAI
SSGITTLVGGGTGPAAGTRATTCTPSPTQMRLMLQSTDYLPLNFGFTGKGSSSKPDELHE
IIKAGAMGLKLHEDWGSTPAAIDNCLTIAEHHDIQINIHTDTLNEAGFVEHSIAAFKGRT
IHTYHSEGAGGGHAPDIIKVCGIKNVLPSSTNPTRPLTSNTIDEHLDMLMVCHHLDREIP
EDLAFAHSRIRKKTIAAEDVLNDIGAISIISSDSQAMGRVGEVISRTWQTADPMKAQTGP
LKCDSSDNDNFRIRRYIAKYTINPAIANGFSQYVGSVEVGKLADLVMWKPSFFGTKPEMV
IKGGMVAWADIGDPNASIPTPEPVKMRPMYGTLGKAGGALSIAFVSKAALDQRVNVLYGL
NKRVEAVSNVRKLTKLDMKLNDALPEITVDPESYTVKADGKLLCVSEATTVPLSRNYFLF
|
|
|
|---|
| BDBM50099857 |
|---|
| n/a |
|---|
| Name | BDBM50099857 |
|---|
| Synonyms: | ACETOHYDROXAMIC ACID (AHA) | AHA | Acethydroxamsaeure | Acethydroxamsaure | Acetic acid, oxime | Acetylhydroxamic acid | Cetohyroxamic acid | Lithostat | Methylhydroxamic acid | N-Acetyl hydroxyacetamide | N-Acetylhydroxylamine | N-Hydroxyacetamide | acetohydroxamic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C2H5NO2 |
|---|
| Mol. Mass. | 75.0666 |
|---|
| SMILES | CC(=O)NO |
|---|
| Structure | 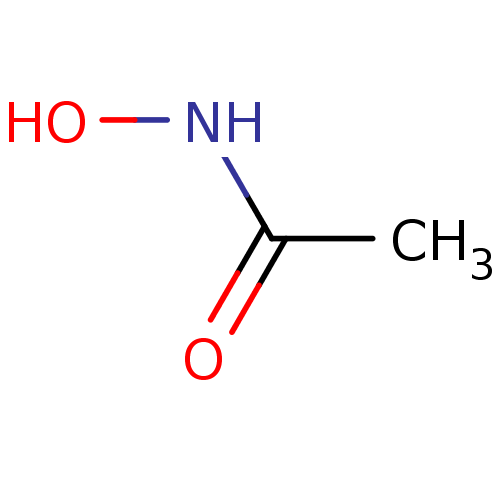
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Saeed, A; Khan, MS; Rafique, H; Shahid, M; Iqbal, J Design, synthesis, molecular docking studies and in vitro screening of ethyl 4-(3-benzoylthioureido) benzoates as urease inhibitors. Bioorg Chem52:1-7 (2014) [PubMed] Article
Saeed, A; Khan, MS; Rafique, H; Shahid, M; Iqbal, J Design, synthesis, molecular docking studies and in vitro screening of ethyl 4-(3-benzoylthioureido) benzoates as urease inhibitors. Bioorg Chem52:1-7 (2014) [PubMed] Article