| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phospholipase D2 |
|---|
| Ligand | BDBM123747 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | PLD In Vitro Kinetic Assay |
|---|
| Temperature | 310.15±0 K |
|---|
| IC50 | 290±n/a nM |
|---|
| Citation |  Kulkarni, A; Quang, P; Curry, V; Keyes, R; Zhou, W; Cho, H; Baffoe, J; Torok, B; Stieglitz, K 1,3-disubstituted-4-aminopyrazolo [3, 4-d] pyrimidines, a new class of potent inhibitors for phospholipase D. Chem Biol Drug Des84:270-81 (2014) [PubMed] Article Kulkarni, A; Quang, P; Curry, V; Keyes, R; Zhou, W; Cho, H; Baffoe, J; Torok, B; Stieglitz, K 1,3-disubstituted-4-aminopyrazolo [3, 4-d] pyrimidines, a new class of potent inhibitors for phospholipase D. Chem Biol Drug Des84:270-81 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Phospholipase D2 |
|---|
| Name: | Phospholipase D2 |
|---|
| Synonyms: | PLD2 | PLD2_HUMAN | Phospholipase D2 | Phospholipase D2 (PLD2) | phospholipase D2 isoform PLD2A |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 106002.47 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 933 |
|---|
| Sequence: | MTATPESLFPTGDELDSSQLQMESDEVDTLKEGEDPADRMHPFLAIYELQSLKVHPLVFA
PGVPVTAQVVGTERYTSGSKVGTCTLYSVRLTHGDFSWTTKKKYRHFQELHRDLLRHKVL
MSLLPLARFAVAYSPARDAGNREMPSLPRAGPEGSTRHAASKQKYLENYLNRLLTMSFYR
NYHAMTEFLEVSQLSFIPDLGRKGLEGMIRKRSGGHRVPGLTCCGRDQVCYRWSKRWLVV
KDSFLLYMCLETGAISFVQLFDPGFEVQVGKRSTEARHGVRIDTSHRSLILKCSSYRQAR
WWAQEITELAQGPGRDFLQLHRHDSYAPPRPGTLARWFVNGAGYFAAVADAILRAQEEIF
ITDWWLSPEVYLKRPAHSDDWRLDIMLKRKAEEGVRVSILLFKEVELALGINSGYSKRAL
MLLHPNIKVMRHPDQVTLWAHHEKLLVVDQVVAFLGGLDLAYGRWDDLHYRLTDLGDSSE
SAASQPPTPRPDSPATPDLSHNQFFWLGKDYSNLITKDWVQLDRPFEDFIDRETTPRMPW
RDVGVVVHGLPARDLARHFIQRWNFTKTTKAKYKTPTYPYLLPKSTSTANQLPFTLPGGQ
CTTVQVLRSVDRWSAGTLENSILNAYLHTIRESQHFLYIENQFFISCSDGRTVLNKVGDE
IVDRILKAHKQGWCYRVYVLLPLLPGFEGDISTGGGNSIQAILHFTYRTLCRGEYSILHR
LKAAMGTAWRDYISICGLRTHGELGGHPVSELIYIHSKVLIADDRTVIIGSANINDRSLL
GKRDSELAVLIEDTETEPSLMNGAEYQAGRFALSLRKHCFGVILGANTRPDLDLRDPICD
DFFQLWQDMAESNANIYEQIFRCLPSNATRSLRTLREYVAVEPLATVSPPLARSELTQVQ
GHLVHFPLKFLEDESLLPPLGSKEGMIPLEVWT
|
|
|
|---|
| BDBM123747 |
|---|
| n/a |
|---|
| Name | BDBM123747 |
|---|
| Synonyms: | 1-(tert-butyl)-3-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine (5) | 1-tert-butyl-3-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine (5) | CHEMBL2069938 | US10544104, Compound 140 | US9765037, Compound 140 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H17N5 |
|---|
| Mol. Mass. | 267.329 |
|---|
| SMILES | CC(C)(C)n1nc(-c2ccccc2)c2c(N)ncnc12 |
|---|
| Structure | 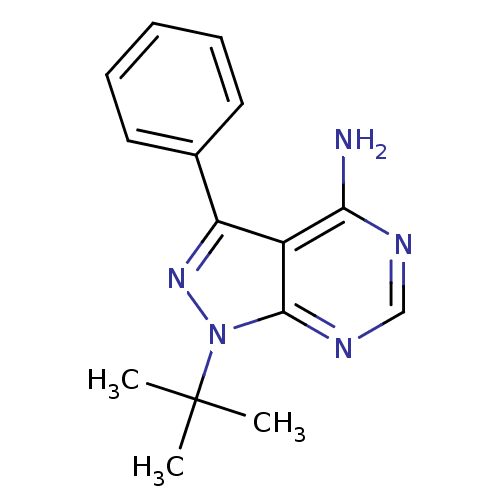
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kulkarni, A; Quang, P; Curry, V; Keyes, R; Zhou, W; Cho, H; Baffoe, J; Torok, B; Stieglitz, K 1,3-disubstituted-4-aminopyrazolo [3, 4-d] pyrimidines, a new class of potent inhibitors for phospholipase D. Chem Biol Drug Des84:270-81 (2014) [PubMed] Article
Kulkarni, A; Quang, P; Curry, V; Keyes, R; Zhou, W; Cho, H; Baffoe, J; Torok, B; Stieglitz, K 1,3-disubstituted-4-aminopyrazolo [3, 4-d] pyrimidines, a new class of potent inhibitors for phospholipase D. Chem Biol Drug Des84:270-81 (2014) [PubMed] Article