| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cGMP-specific 3',5'-cyclic phosphodiesterase |
|---|
| Ligand | BDBM131007 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Inhibition Assay |
|---|
| Temperature | 303.15±n/a K |
|---|
| IC50 | >10000±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Okada, M; Sato, S; Kawade, K; Gotanda, K; Shinbo, A; Nakano, Y; Kobayashi, H Substituted imidazo[1,5-A]quinoxalines as phosphodiesterase 9 inhibitors US Patent US8829000 Publication Date 9/9/2014 Okada, M; Sato, S; Kawade, K; Gotanda, K; Shinbo, A; Nakano, Y; Kobayashi, H Substituted imidazo[1,5-A]quinoxalines as phosphodiesterase 9 inhibitors US Patent US8829000 Publication Date 9/9/2014 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cGMP-specific 3',5'-cyclic phosphodiesterase |
|---|
| Name: | cGMP-specific 3',5'-cyclic phosphodiesterase |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | CGB-PDE | PDE5 | PDE5A | PDE5A_HUMAN | Phosphodiesterase 2 and 5 (PDE2 and PDE5) | Phosphodiesterase 5 (PDE5) | Phosphodiesterase 5A | Phosphodiesterase 5A (PDE5A) | cGMP-binding cGMP-specific phosphodiesterase | cGMP-specific 3',5'-cyclic phosphodiesterase |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 99975.83 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O76074 |
|---|
| Residue: | 875 |
|---|
| Sequence: | MERAGPSFGQQRQQQQPQQQKQQQRDQDSVEAWLDDHWDFTFSYFVRKATREMVNAWFAE
RVHTIPVCKEGIRGHTESCSCPLQQSPRADNSAPGTPTRKISASEFDRPLRPIVVKDSEG
TVSFLSDSEKKEQMPLTPPRFDHDEGDQCSRLLELVKDISSHLDVTALCHKIFLHIHGLI
SADRYSLFLVCEDSSNDKFLISRLFDVAEGSTLEEVSNNCIRLEWNKGIVGHVAALGEPL
NIKDAYEDPRFNAEVDQITGYKTQSILCMPIKNHREEVVGVAQAINKKSGNGGTFTEKDE
KDFAAYLAFCGIVLHNAQLYETSLLENKRNQVLLDLASLIFEEQQSLEVILKKIAATIIS
FMQVQKCTIFIVDEDCSDSFSSVFHMECEELEKSSDTLTREHDANKINYMYAQYVKNTME
PLNIPDVSKDKRFPWTTENTGNVNQQCIRSLLCTPIKNGKKNKVIGVCQLVNKMEENTGK
VKPFNRNDEQFLEAFVIFCGLGIQNTQMYEAVERAMAKQMVTLEVLSYHASAAEEETREL
QSLAAAVVPSAQTLKITDFSFSDFELSDLETALCTIRMFTDLNLVQNFQMKHEVLCRWIL
SVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLEILALLIAALSHDLDHRGV
NNSYIQRSEHPLAQLYCHSIMEHHHFDQCLMILNSPGNQILSGLSIEEYKTTLKIIKQAI
LATDLALYIKRRGEFFELIRKNQFNLEDPHQKELFLAMLMTACDLSAITKPWPIQQRIAE
LVATEFFDQGDRERKELNIEPTDLMNREKKNKIPSMQVGFIDAICLQLYEALTHVSEDCF
PLLDGCRKNRQKWQALAEQQEKMLINGESGQAKRN
|
|
|
|---|
| BDBM131007 |
|---|
| n/a |
|---|
| Name | BDBM131007 |
|---|
| Synonyms: | US8829000, 9 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H18N4O2 |
|---|
| Mol. Mass. | 298.3397 |
|---|
| SMILES | CC(C)c1ncc2n1c1ccc(cc1[nH]c2=O)C(=O)N(C)C |
|---|
| Structure | 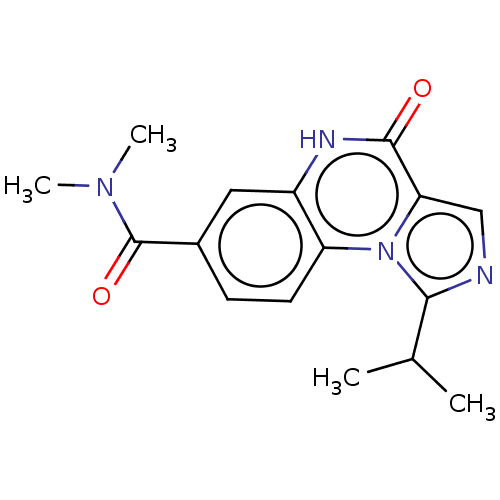
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Okada, M; Sato, S; Kawade, K; Gotanda, K; Shinbo, A; Nakano, Y; Kobayashi, H Substituted imidazo[1,5-A]quinoxalines as phosphodiesterase 9 inhibitors US Patent US8829000 Publication Date 9/9/2014
Okada, M; Sato, S; Kawade, K; Gotanda, K; Shinbo, A; Nakano, Y; Kobayashi, H Substituted imidazo[1,5-A]quinoxalines as phosphodiesterase 9 inhibitors US Patent US8829000 Publication Date 9/9/2014