

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Isoform 4 of cAMP-specific 3',5'-cyclic phosphodiesterase 4A (RD1) | ||
| Ligand | BDBM285626 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | The PDE4A3, PDE4B1, PDE4C1 and PDE4D3 assays | ||
| IC50 | 4.58±n/a nM | ||
| Citation |  Chappie, TA; Patel, NC; Hayward, MM; Helal, CJ; Sciabola, S; LaChapelle, EA; Young, JM; Verhoest, PR Imidazopyridazine compounds US Patent US10077269 Publication Date 9/18/2018 Chappie, TA; Patel, NC; Hayward, MM; Helal, CJ; Sciabola, S; LaChapelle, EA; Young, JM; Verhoest, PR Imidazopyridazine compounds US Patent US10077269 Publication Date 9/18/2018 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Isoform 4 of cAMP-specific 3',5'-cyclic phosphodiesterase 4A (RD1) | |||
| Name: | Isoform 4 of cAMP-specific 3',5'-cyclic phosphodiesterase 4A (RD1) | ||
| Synonyms: | DPDE2 | PDE4A | PDE4A1 | PDE4A_HUMAN | Phosphodiesterase 4A (PDE4A1) | Phosphodiesterase 4A1 | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (RD1) | cAMP-specific 3',5'-cyclic phosphodiesterase 4A Isoform 4 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 72186.86 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P27815-4 | ||
| Residue: | 647 | ||
| Sequence: |
| ||
| BDBM285626 | |||
| n/a | |||
| Name | BDBM285626 | ||
| Synonyms: | 3-(4-chloro-2,5-difluoro-phenyl)-N-ethylimidazo[1,2-b]pyridazine-2-carboxamide | US10077269, Example 96 | US10669279, Example 96 | US9598421, Example 96 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C15H11ClF2N4O | ||
| Mol. Mass. | 336.724 | ||
| SMILES | CCNC(=O)c1nc2cccnn2c1-c1cc(F)c(Cl)cc1F |(4.83,-4.93,;4.06,-3.6,;2.52,-3.6,;1.75,-2.26,;2.52,-.93,;.21,-2.26,;-.7,-3.51,;-2.16,-3.03,;-3.49,-3.8,;-4.83,-3.03,;-4.83,-1.49,;-3.49,-.72,;-2.16,-1.49,;-.7,-1.02,;-.3,.47,;1.19,.87,;1.59,2.36,;3.08,2.75,;.5,3.44,;.9,4.93,;-.99,3.05,;-1.39,1.56,;-2.87,1.16,)| | ||
| Structure | 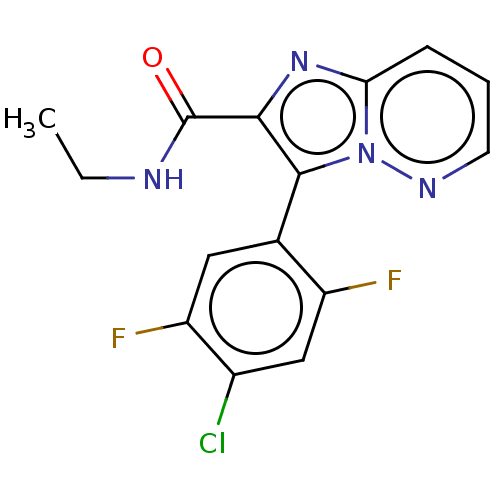
| ||