| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Beta-glucuronidase |
|---|
| Ligand | BDBM163645 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Inhibition Assays |
|---|
| Ki | 6.1e+2± 2e+2 nM |
|---|
| Citation |  Wallace, BD; Roberts, AB; Pollet, RM; Ingle, JD; Biernat, KA; Pellock, SJ; Venkatesh, MK; Guthrie, L; O'Neal, SK; Robinson, SJ; Dollinger, M; Figueroa, E; McShane, SR; Cohen, RD; Jin, J; Frye, SV; Zamboni, WC; Pepe-Ranney, C; Mani, S; Kelly, L; Redinbo, MR Structure and Inhibition of Microbiome ß-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem Biol22:1238-49 (2015) [PubMed] Article Wallace, BD; Roberts, AB; Pollet, RM; Ingle, JD; Biernat, KA; Pellock, SJ; Venkatesh, MK; Guthrie, L; O'Neal, SK; Robinson, SJ; Dollinger, M; Figueroa, E; McShane, SR; Cohen, RD; Jin, J; Frye, SV; Zamboni, WC; Pepe-Ranney, C; Mani, S; Kelly, L; Redinbo, MR Structure and Inhibition of Microbiome ß-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem Biol22:1238-49 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Beta-glucuronidase |
|---|
| Name: | Beta-glucuronidase |
|---|
| Synonyms: | β-Glucuronidase | BGLR_ECOLI | Beta-glucuronidase (EcGUS) | gurA | gusA | uidA |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 68431.57 |
|---|
| Organism: | Escherichia coli (Enterobacteria) |
|---|
| Description: | P05804 |
|---|
| Residue: | 603 |
|---|
| Sequence: | MLRPVETPTREIKKLDGLWAFSLDRENCGIDQRWWESALQESRAIAVPGSFNDQFADADI
RNYAGNVWYQREVFIPKGWAGQRIVLRFDAVTHYGKVWVNNQEVMEHQGGYTPFEADVTP
YVIAGKSVRITVCVNNELNWQTIPPGMVITDENGKKKQSYFHDFFNYAGIHRSVMLYTTP
NTWVDDITVVTHVAQDCNHASVDWQVVANGDVSVELRDADQQVVATGQGTSGTLQVVNPH
LWQPGEGYLYELCVTAKSQTECDIYPLRVGIRSVAVKGEQFLINHKPFYFTGFGRHEDAD
LRGKGFDNVLMVHDHALMDWIGANSYRTSHYPYAEEMLDWADEHGIVVIDETAAVGFNLS
LGIGFEAGNKPKELYSEEAVNGETQQAHLQAIKELIARDKNHPSVVMWSIANEPDTRPQG
AREYFAPLAEATRKLDPTRPITCVNVMFCDAHTDTISDLFDVLCLNRYYGWYVQSGDLET
AEKVLEKELLAWQEKLHQPIIITEYGVDTLAGLHSMYTDMWSEEYQCAWLDMYHRVFDRV
SAVVGEQVWNFADFATSQGILRVGGNKKGIFTRDRKPKSAAFLLQKRWTGMNFGEKPQQG
GKQ
|
|
|
|---|
| BDBM163645 |
|---|
| n/a |
|---|
| Name | BDBM163645 |
|---|
| Synonyms: | 3[(6,8Dimethyl2oxo1Hquinolin3yl)methyl]3(2hydroxyethyl)1[4(morpholin4yl)phenyl]thiourea (Inhibitor R3) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H30N4O3S |
|---|
| Mol. Mass. | 466.596 |
|---|
| SMILES | Cc1cc(C)c2[nH]c(=O)c(CN(CCO)C(=S)Nc3ccc(cc3)N3CCOCC3)cc2c1 |
|---|
| Structure | 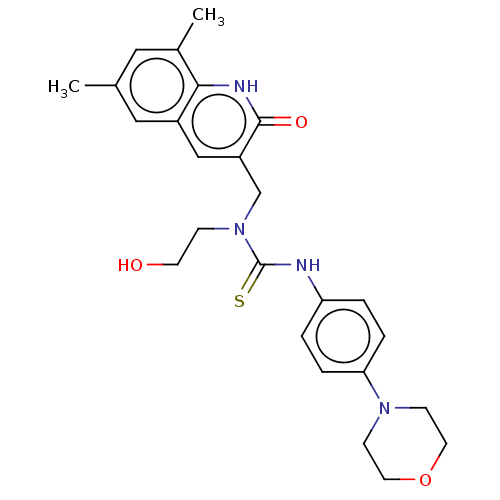
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wallace, BD; Roberts, AB; Pollet, RM; Ingle, JD; Biernat, KA; Pellock, SJ; Venkatesh, MK; Guthrie, L; O'Neal, SK; Robinson, SJ; Dollinger, M; Figueroa, E; McShane, SR; Cohen, RD; Jin, J; Frye, SV; Zamboni, WC; Pepe-Ranney, C; Mani, S; Kelly, L; Redinbo, MR Structure and Inhibition of Microbiome ß-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem Biol22:1238-49 (2015) [PubMed] Article
Wallace, BD; Roberts, AB; Pollet, RM; Ingle, JD; Biernat, KA; Pellock, SJ; Venkatesh, MK; Guthrie, L; O'Neal, SK; Robinson, SJ; Dollinger, M; Figueroa, E; McShane, SR; Cohen, RD; Jin, J; Frye, SV; Zamboni, WC; Pepe-Ranney, C; Mani, S; Kelly, L; Redinbo, MR Structure and Inhibition of Microbiome ß-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem Biol22:1238-49 (2015) [PubMed] Article