| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Neuraminidase |
|---|
| Ligand | BDBM5202 |
|---|
| Substrate/Competitor | BDBM4702 |
|---|
| Meas. Tech. | Neuraminidase Inhibition Assay |
|---|
| Ki | 20±n/a nM |
|---|
| Citation |  Maring, CJ; Stoll, VS; Zhao, C; Sun, M; Krueger, AC; Stewart, KD; Madigan, DL; Kati, WM; Xu, Y; Carrick, RJ; Montgomery, DA; Kempf-Grote, A; Marsh, KC; Molla, A; Steffy, KR; Sham, HL; Laver, WG; Gu, YG; Kempf, DJ; Kohlbrenner, WE Structure-based characterization and optimization of novel hydrophobic binding interactions in a series of pyrrolidine influenza neuraminidase inhibitors. J Med Chem48:3980-90 (2005) [PubMed] Article Maring, CJ; Stoll, VS; Zhao, C; Sun, M; Krueger, AC; Stewart, KD; Madigan, DL; Kati, WM; Xu, Y; Carrick, RJ; Montgomery, DA; Kempf-Grote, A; Marsh, KC; Molla, A; Steffy, KR; Sham, HL; Laver, WG; Gu, YG; Kempf, DJ; Kohlbrenner, WE Structure-based characterization and optimization of novel hydrophobic binding interactions in a series of pyrrolidine influenza neuraminidase inhibitors. J Med Chem48:3980-90 (2005) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Neuraminidase |
|---|
| Name: | Neuraminidase |
|---|
| Synonyms: | Influenza A Virus Neuraminidase | NA | NRAM_I67A0 | Neuraminidase A |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52132.25 |
|---|
| Organism: | Influenza A virus (A/Tokyo/3/67(H2N2)) |
|---|
| Description: | P06820 |
|---|
| Residue: | 469 |
|---|
| Sequence: | MNPNQKIITIGSVSLTIATVCFLMQIAILVTTVTLHFKQHECDSPASNQVMPCEPIIIER
NITEIVYLNNTTIEKEICPKVVEYRNWSKPQCQITGFAPFSKDNSIRLSAGGDIWVTREP
YVSCDPVKCYQFALGQGTTLDNKHSNDTVHDRIPHRTLLMNELGVPFHLGTRQVCIAWSS
SSCHDGKAWLHVCITGDDKNATASFIYDGRLVDSIGSWSQNILRTQESECVCINGTCTVV
MTDGSASGRADTRILFIEEGKIVHISPLAGSAQHVEECSCYPRYPGVRCICRDNWKGSNR
PVVDINMEDYSIDSSYVCSGLVGDTPRNDDRSSNSNCRNPNNERGTQGVKGWAFDNGNDL
WMGRTISKDLRSGYETFKVIGGWSTPNSKSQINRQVIVDSDNRSGYSGIFSVEGKSCINR
CFYVELIRGRKQETRVWWTSNSIVVFCGTSGTYGTGSWPDGANINFMPI
|
|
|
|---|
| BDBM5202 |
|---|
| BDBM4702 |
|---|
| Name | BDBM5202 |
|---|
| Synonyms: | (+/-)-(2R,4S,5R,1 S)-5-(1 -Acetylamino-3 -methyl-butyl)-4-(trans-propen-1-yl)-pyrrolidine-2-carboxylic acid hydrochloric acid salt | (2R,4S,5R)-5-[(1S)-1-acetamido-3-methylbutyl]-4-[(1E)-prop-1-en-1-yl]pyrrolidine-2-carboxylic acid | Pyrrolidine deriv. 30 | Pyrrolidine deriv. 32 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H26N2O3 |
|---|
| Mol. Mass. | 282.3785 |
|---|
| SMILES | [H][C@]1(N[C@H](C[C@H]1\C=C/C)C(O)=O)[C@H](CC(C)C)NC(C)=O |r| |
|---|
| Structure | 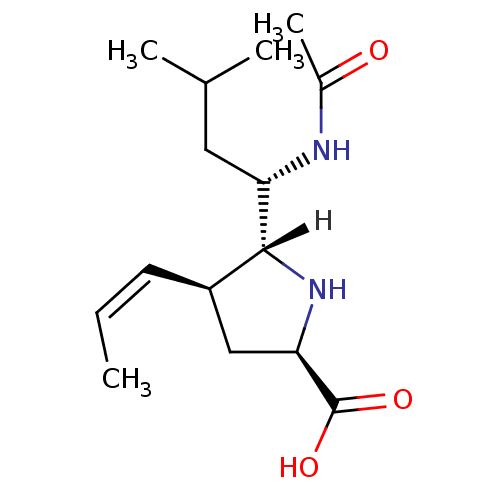
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Maring, CJ; Stoll, VS; Zhao, C; Sun, M; Krueger, AC; Stewart, KD; Madigan, DL; Kati, WM; Xu, Y; Carrick, RJ; Montgomery, DA; Kempf-Grote, A; Marsh, KC; Molla, A; Steffy, KR; Sham, HL; Laver, WG; Gu, YG; Kempf, DJ; Kohlbrenner, WE Structure-based characterization and optimization of novel hydrophobic binding interactions in a series of pyrrolidine influenza neuraminidase inhibitors. J Med Chem48:3980-90 (2005) [PubMed] Article
Maring, CJ; Stoll, VS; Zhao, C; Sun, M; Krueger, AC; Stewart, KD; Madigan, DL; Kati, WM; Xu, Y; Carrick, RJ; Montgomery, DA; Kempf-Grote, A; Marsh, KC; Molla, A; Steffy, KR; Sham, HL; Laver, WG; Gu, YG; Kempf, DJ; Kohlbrenner, WE Structure-based characterization and optimization of novel hydrophobic binding interactions in a series of pyrrolidine influenza neuraminidase inhibitors. J Med Chem48:3980-90 (2005) [PubMed] Article