| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phospholipase D |
|---|
| Ligand | BDBM123753 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | pH-Stat Assay |
|---|
| pH | 8±n/a |
|---|
| IC50 | 900±0.0 nM |
|---|
| Comments | extracted |
|---|
| Citation |  Djakpa, H; Kulkarni, A; Barrows-Murphy, S; Miller, G; Zhou, W; Cho, H; Török, B; Stieglitz, K Identifying New Drug Targets for Potent Phospholipase D Inhibitors: Combining Sequence Alignment, Molecular Docking, and Enzyme Activity/Binding Assays. Chem Biol Drug Des87:714-29 (2016) [PubMed] Article Djakpa, H; Kulkarni, A; Barrows-Murphy, S; Miller, G; Zhou, W; Cho, H; Török, B; Stieglitz, K Identifying New Drug Targets for Potent Phospholipase D Inhibitors: Combining Sequence Alignment, Molecular Docking, and Enzyme Activity/Binding Assays. Chem Biol Drug Des87:714-29 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Phospholipase D |
|---|
| Name: | Phospholipase D |
|---|
| Synonyms: | PLD_STRCW | Phospholipase D (PLD_SC) | pld |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 59647.22 |
|---|
| Organism: | Streptomyces chromofuscus |
|---|
| Description: | Q8KRU5 |
|---|
| Residue: | 556 |
|---|
| Sequence: | MTSRYRSSEAHQGLASFSPRRRTVVKAAAATAVLAGPLAAALPARATTGTPAFLHGVASG
DPLPDGVLLWTRVTPTADATPGSGLGPDTEVGWTVATDKAFTNVVAKGSTTATAASDHTV
KADIRGLAPATDHWFRFSAGGTDSPAGRARTAPAADAAVAGLRFGVVSCANWEAGYFAAY
RHLAARGDLDAWLHLGDYIYEYGAGEYGTRGTSVRSHAPAHEILTLADYRVRHGRYKTDP
DLQALHAAAPVVAIWDDHEIANDTWSGGAENHTEGVEGAWAARQAAAKQAYFEWMPVRPA
IAGTTYRRLRFGKLADLSLLDLRSFRAQQVSLGDGDVDDPDRTLTGRAQLDWLKAGLKSS
DTTWRLVGNSVMIAPFAIGSLSAELLKPLAKLLGLPQEGLAVNTDQWDGYTDDRRELLAH
LRSNAIRNTVFLTGDIHMAWANDVPVNAGTYPLSASAATEFVVTSVTSDNLDDLVKVPEG
TVSALASPVIRAANRHVHWVDTDRHGYGVLDITAERAQMDYYVLSDRTQAGATASWSRSY
RTRSGTQRVERTYDPE
|
|
|
|---|
| BDBM123753 |
|---|
| n/a |
|---|
| Name | BDBM123753 |
|---|
| Synonyms: | 3-methyl-1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine (12) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H11N5 |
|---|
| Mol. Mass. | 225.2492 |
|---|
| SMILES | Cc1nn(-c2ccccc2)c2ncnc(N)c12 |
|---|
| Structure | 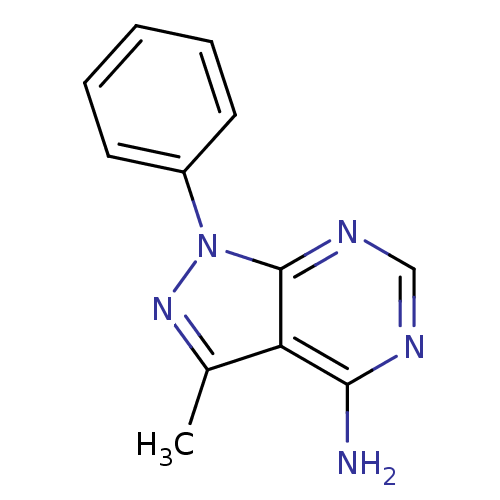
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Djakpa, H; Kulkarni, A; Barrows-Murphy, S; Miller, G; Zhou, W; Cho, H; Török, B; Stieglitz, K Identifying New Drug Targets for Potent Phospholipase D Inhibitors: Combining Sequence Alignment, Molecular Docking, and Enzyme Activity/Binding Assays. Chem Biol Drug Des87:714-29 (2016) [PubMed] Article
Djakpa, H; Kulkarni, A; Barrows-Murphy, S; Miller, G; Zhou, W; Cho, H; Török, B; Stieglitz, K Identifying New Drug Targets for Potent Phospholipase D Inhibitors: Combining Sequence Alignment, Molecular Docking, and Enzyme Activity/Binding Assays. Chem Biol Drug Des87:714-29 (2016) [PubMed] Article