| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 3 |
|---|
| Ligand | BDBM49152 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | HDAC Enzymatic Assay |
|---|
| pH | 7.4±n/a |
|---|
| IC50 | 6.51e+4±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Boskovic, ZV; Kemp, MM; Freedy, AM; Viswanathan, VS; Pop, MS; Fuller, JH; Martinez, NM; Figueroa Laz˙, SO; Hong, JA; Lewis, TA; Calarese, D; Love, JD; Vetere, A; Almo, SC; Schreiber, SL; Koehler, AN Inhibition of Zinc-Dependent Histone Deacetylases with a Chemically Triggered Electrophile. ACS Chem Biol11:1844-51 (2016) [PubMed] Article Boskovic, ZV; Kemp, MM; Freedy, AM; Viswanathan, VS; Pop, MS; Fuller, JH; Martinez, NM; Figueroa Laz˙, SO; Hong, JA; Lewis, TA; Calarese, D; Love, JD; Vetere, A; Almo, SC; Schreiber, SL; Koehler, AN Inhibition of Zinc-Dependent Histone Deacetylases with a Chemically Triggered Electrophile. ACS Chem Biol11:1844-51 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 3 |
|---|
| Name: | Histone deacetylase 3 |
|---|
| Synonyms: | HD3 | HDAC3 | HDAC3_HUMAN | Histone deacetylase 3 (HDAC3) | Human HDAC3 | RPD3-2 | SMAP45 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48829.55 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15379 |
|---|
| Residue: | 428 |
|---|
| Sequence: | MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN
KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA
FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI
NQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV
RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ
TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN
DKESDVEI
|
|
|
|---|
| BDBM49152 |
|---|
| n/a |
|---|
| Name | BDBM49152 |
|---|
| Synonyms: | 5-chloranyl-7-[(4-ethylpiperazin-1-yl)-pyridin-3-yl-methyl]quinolin-8-ol | 5-chloro-7-[(4-ethyl-1-piperazinyl)-(3-pyridinyl)methyl]-8-quinolinol | 5-chloro-7-[(4-ethylpiperazin-1-yl)-pyridin-3-ylmethyl]quinolin-8-ol | 5-chloro-7-[(4-ethylpiperazino)-(3-pyridyl)methyl]quinolin-8-ol | BRD4354 | MLS000564806 | SMR000151966 | cid_3516032 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H23ClN4O |
|---|
| Mol. Mass. | 382.887 |
|---|
| SMILES | CCN1CCN(CC1)C(c1cccnc1)c1cc(Cl)c2cccnc2c1O |
|---|
| Structure | 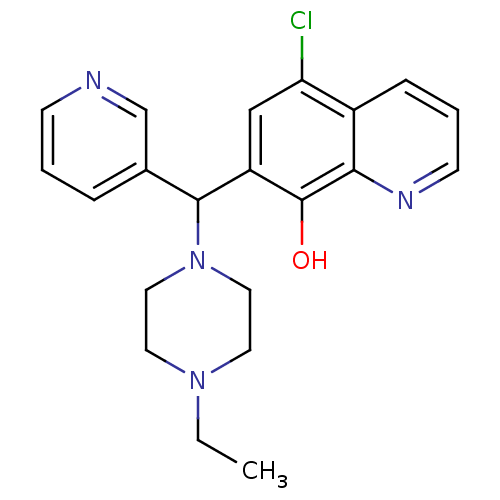
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Boskovic, ZV; Kemp, MM; Freedy, AM; Viswanathan, VS; Pop, MS; Fuller, JH; Martinez, NM; Figueroa Laz˙, SO; Hong, JA; Lewis, TA; Calarese, D; Love, JD; Vetere, A; Almo, SC; Schreiber, SL; Koehler, AN Inhibition of Zinc-Dependent Histone Deacetylases with a Chemically Triggered Electrophile. ACS Chem Biol11:1844-51 (2016) [PubMed] Article
Boskovic, ZV; Kemp, MM; Freedy, AM; Viswanathan, VS; Pop, MS; Fuller, JH; Martinez, NM; Figueroa Laz˙, SO; Hong, JA; Lewis, TA; Calarese, D; Love, JD; Vetere, A; Almo, SC; Schreiber, SL; Koehler, AN Inhibition of Zinc-Dependent Histone Deacetylases with a Chemically Triggered Electrophile. ACS Chem Biol11:1844-51 (2016) [PubMed] Article