| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosyl-DNA phosphodiesterase 2 [E242G,S244A,Q278R,I281T,K282R,R316G,P318T,Y321CH323L] |
|---|
| Ligand | BDBM50438865 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Recombinant TDP2 Assay |
|---|
| pH | 7.5±n/a |
|---|
| IC50 | 260±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Marchand, C; Abdelmalak, M; Kankanala, J; Huang, SY; Kiselev, E; Fesen, K; Kurahashi, K; Sasanuma, H; Takeda, S; Aihara, H; Wang, Z; Pommier, Y Deazaflavin Inhibitors of Tyrosyl-DNA Phosphodiesterase 2 (TDP2) Specific for the Human Enzyme and Active against Cellular TDP2. ACS Chem Biol11:1925-33 (2016) [PubMed] Article Marchand, C; Abdelmalak, M; Kankanala, J; Huang, SY; Kiselev, E; Fesen, K; Kurahashi, K; Sasanuma, H; Takeda, S; Aihara, H; Wang, Z; Pommier, Y Deazaflavin Inhibitors of Tyrosyl-DNA Phosphodiesterase 2 (TDP2) Specific for the Human Enzyme and Active against Cellular TDP2. ACS Chem Biol11:1925-33 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosyl-DNA phosphodiesterase 2 [E242G,S244A,Q278R,I281T,K282R,R316G,P318T,Y321CH323L] |
|---|
| Name: | Tyrosyl-DNA phosphodiesterase 2 [E242G,S244A,Q278R,I281T,K282R,R316G,P318T,Y321CH323L] |
|---|
| Synonyms: | TYDP2_MOUSE | Tdp2 | Ttrap | Tyrosyl-DNA phosphodiesterase 2 mutant 9M(mTDP2 9M) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 40804.32 |
|---|
| Organism: | Mus musculus (Mouse) |
|---|
| Description: | The mTDP2 9M bears the mutations at residues Q278R, I281T, K282R, P318T, Y321C, E242G, S244A, R316G, and H323L. |
|---|
| Residue: | 370 |
|---|
| Sequence: | MASGSSSDAAEPAGPAGRAASAPEAAQAEEDRVKRRRLQCLGFALVGGCDPTMVPSVLRE
NDWQTQKALSAYFELPENDQGWPRQPPTSFKSEAYVDLTNEDANDTTILEASPSGTPLED
SSTISFITWNIDGLDGCNLPERARGVCSCLALYSPDVVFLQEVIPPYCAYLKKRAASYTI
ITGNEEGYFTAILLKKGRVKFKSQEIIPFPNTKMMRNLLCVNVSLGGNEFCLMTSHLEST
RGHAAERIRQLKTVLGKMQEAPDSTTVIFAGDTNLRDREVTRCGGLPDNVFDAWEFLGKP
KHCQYTWDTKANNNLGITAACKLRFDRIFFRAEEGHLIPQSLDLVGLEKLDCGRFPSDHW
GLLCTLNVVL
|
|
|
|---|
| BDBM50438865 |
|---|
| n/a |
|---|
| Name | BDBM50438865 |
|---|
| Synonyms: | 2,4-Dioxo-10-phenyl-pyrimido[4,5-b]quinoline-8-carbonitrile (3) | CHEMBL2420506 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H10N4O2 |
|---|
| Mol. Mass. | 314.2976 |
|---|
| SMILES | O=c1nc2n(-c3ccccc3)c3cc(ccc3cc2c(=O)[nH]1)C#N |
|---|
| Structure | 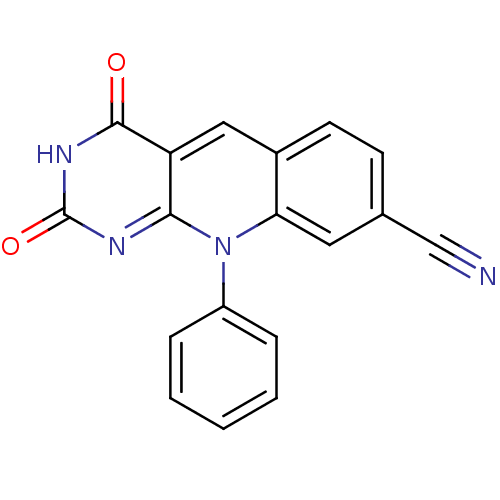
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Marchand, C; Abdelmalak, M; Kankanala, J; Huang, SY; Kiselev, E; Fesen, K; Kurahashi, K; Sasanuma, H; Takeda, S; Aihara, H; Wang, Z; Pommier, Y Deazaflavin Inhibitors of Tyrosyl-DNA Phosphodiesterase 2 (TDP2) Specific for the Human Enzyme and Active against Cellular TDP2. ACS Chem Biol11:1925-33 (2016) [PubMed] Article
Marchand, C; Abdelmalak, M; Kankanala, J; Huang, SY; Kiselev, E; Fesen, K; Kurahashi, K; Sasanuma, H; Takeda, S; Aihara, H; Wang, Z; Pommier, Y Deazaflavin Inhibitors of Tyrosyl-DNA Phosphodiesterase 2 (TDP2) Specific for the Human Enzyme and Active against Cellular TDP2. ACS Chem Biol11:1925-33 (2016) [PubMed] Article