

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Histone-lysine N-methyltransferase EZH2 [Y641F] | ||
| Ligand | BDBM190217 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Inhibition Assay | ||
| IC50 | 3.1±n/a nM | ||
| Citation |  Kuntz, KW; Knutson, SK; Wigle, TJ Inhibitors of human EZH2, and methods of use thereof US Patent US9175331 Publication Date 11/3/2015 Kuntz, KW; Knutson, SK; Wigle, TJ Inhibitors of human EZH2, and methods of use thereof US Patent US9175331 Publication Date 11/3/2015 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Histone-lysine N-methyltransferase EZH2 [Y641F] | |||
| Name: | Histone-lysine N-methyltransferase EZH2 [Y641F] | ||
| Synonyms: | EZH2 | EZH2(Y641F) | EZH2_HUMAN | Histone-lysine N-methyltransferase EZH2 (Y641F) | KMT6 | ||
| Type: | n/a | ||
| Mol. Mass.: | 85351.84 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q15910[Y641F] | ||
| Residue: | 746 | ||
| Sequence: |
| ||
| BDBM190217 | |||
| n/a | |||
| Name | BDBM190217 | ||
| Synonyms: | EPZ009152 | US9175331, 54 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H36ClN3O4 | ||
| Mol. Mass. | 502.045 | ||
| SMILES | CCOC(=O)[C@H]1CC[C@@H](CC1)N(CC)c1cc(Cl)cc(C(=O)NCc2c(C)cc(C)[nH]c2=O)c1C |r,wU:5.4,wD:8.11,(6.67,8.47,;5.33,7.7,;4,8.47,;2.67,7.7,;1.33,8.47,;2.67,6.16,;4,5.39,;4,3.85,;2.67,3.08,;1.33,3.85,;1.33,5.39,;2.67,1.54,;4,.77,;4,-.77,;1.33,.77,;,1.54,;-1.33,.77,;-2.67,1.54,;-1.33,-.77,;,-1.54,;,-3.08,;1.33,-3.85,;-1.33,-3.85,;-1.33,-5.39,;-2.67,-6.16,;-2.67,-7.7,;-1.33,-8.47,;-4,-8.47,;-5.33,-7.7,;-6.67,-8.47,;-5.33,-6.16,;-4,-5.39,;-4,-3.85,;1.33,-.77,;2.67,-1.54,)| | ||
| Structure | 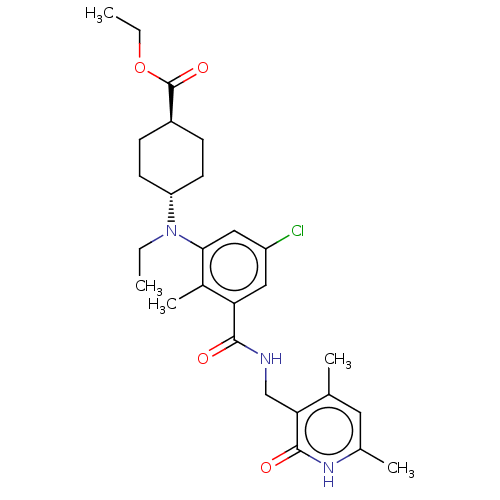
| ||