| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Isocitrate dehydrogenase [NADP], mitochondrial [R172S] |
|---|
| Ligand | BDBM195602 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | RapidFire-MS/MS and NADPH Assays |
|---|
| Temperature | 298.15±n/a K |
|---|
| IC50 | >100000±0.0 nM |
|---|
| Comments | extracted |
|---|
| Citation |  Okoye-Okafor, UC; Bartholdy, B; Cartier, J; Gao, EN; Pietrak, B; Rendina, AR; Rominger, C; Quinn, C; Smallwood, A; Wiggall, KJ; Reif, AJ; Schmidt, SJ; Qi, H; Zhao, H; Joberty, G; Faelth-Savitski, M; Bantscheff, M; Drewes, G; Duraiswami, C; Brady, P; Groy, A; Narayanagari, SR; Antony-Debre, I; Mitchell, K; Wang, HR; Kao, YR; Christopeit, M; Carvajal, L; Barreyro, L; Paietta, E; Makishima, H; Will, B; Concha, N; Adams, ND; Schwartz, B; McCabe, MT; Maciejewski, J; Verma, A; Steidl, U New IDH1 mutant inhibitors for treatment of acute myeloid leukemia Nat Chem Biol11:878-86 (2015) [PubMed] Article Okoye-Okafor, UC; Bartholdy, B; Cartier, J; Gao, EN; Pietrak, B; Rendina, AR; Rominger, C; Quinn, C; Smallwood, A; Wiggall, KJ; Reif, AJ; Schmidt, SJ; Qi, H; Zhao, H; Joberty, G; Faelth-Savitski, M; Bantscheff, M; Drewes, G; Duraiswami, C; Brady, P; Groy, A; Narayanagari, SR; Antony-Debre, I; Mitchell, K; Wang, HR; Kao, YR; Christopeit, M; Carvajal, L; Barreyro, L; Paietta, E; Makishima, H; Will, B; Concha, N; Adams, ND; Schwartz, B; McCabe, MT; Maciejewski, J; Verma, A; Steidl, U New IDH1 mutant inhibitors for treatment of acute myeloid leukemia Nat Chem Biol11:878-86 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Isocitrate dehydrogenase [NADP], mitochondrial [R172S] |
|---|
| Name: | Isocitrate dehydrogenase [NADP], mitochondrial [R172S] |
|---|
| Synonyms: | IDH2 | IDHP_HUMAN | Isocitrate dehydrogenase 2 mutant (R172S) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 50853.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | R172S mutant |
|---|
| Residue: | 452 |
|---|
| Sequence: | MAGYLRVVRSLCRASGSRPAWAPAALTAPTSQEQPRRHYADKRIKVAKPVVEMDGDEMTR
IIWQFIKEKLILPHVDIQLKYFDLGLPNRDQTDDQVTIDSALATQKYSVAVKCATITPDE
ARVEEFKLKKMWKSPNGTIRNILGGTVFREPIICKNIPRLVPGWTKPITIGSHAHGDQYK
ATDFVADRAGTFKMVFTPKDGSGVKEWEVYNFPAGGVGMGMYNTDESISGFAHSCFQYAI
QKKWPLYMSTKNTILKAYDGRFKDIFQEIFDKHYKTDFDKNKIWYEHRLIDDMVAQVLKS
SGGFVWACKNYDGDVQSDILAQGFGSLGLMTSVLVCPDGKTIEAEAAHGTVTRHYREHQK
GRPTSTNPIASIFAWTRGLEHRGKLDGNQDLIRFAQMLEKVCVETVESGAMTKDLAGCIH
GLSNVKLNEHFLNTTDFLDTIKSNLDRALGRQ
|
|
|
|---|
| BDBM195602 |
|---|
| n/a |
|---|
| Name | BDBM195602 |
|---|
| Synonyms: | GSK990 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H23N7O2 |
|---|
| Mol. Mass. | 429.4744 |
|---|
| SMILES | Cc1cccc(NC(=O)c2nn(Cc3c[nH]cn3)c3CCN(Cc23)C(=O)c2ccc[nH]2)c1 |
|---|
| Structure | 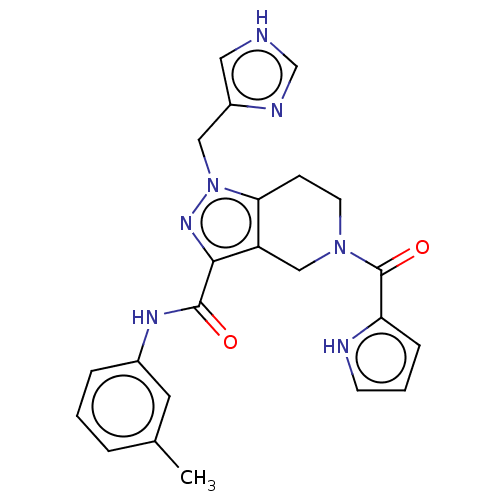
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Okoye-Okafor, UC; Bartholdy, B; Cartier, J; Gao, EN; Pietrak, B; Rendina, AR; Rominger, C; Quinn, C; Smallwood, A; Wiggall, KJ; Reif, AJ; Schmidt, SJ; Qi, H; Zhao, H; Joberty, G; Faelth-Savitski, M; Bantscheff, M; Drewes, G; Duraiswami, C; Brady, P; Groy, A; Narayanagari, SR; Antony-Debre, I; Mitchell, K; Wang, HR; Kao, YR; Christopeit, M; Carvajal, L; Barreyro, L; Paietta, E; Makishima, H; Will, B; Concha, N; Adams, ND; Schwartz, B; McCabe, MT; Maciejewski, J; Verma, A; Steidl, U New IDH1 mutant inhibitors for treatment of acute myeloid leukemia Nat Chem Biol11:878-86 (2015) [PubMed] Article
Okoye-Okafor, UC; Bartholdy, B; Cartier, J; Gao, EN; Pietrak, B; Rendina, AR; Rominger, C; Quinn, C; Smallwood, A; Wiggall, KJ; Reif, AJ; Schmidt, SJ; Qi, H; Zhao, H; Joberty, G; Faelth-Savitski, M; Bantscheff, M; Drewes, G; Duraiswami, C; Brady, P; Groy, A; Narayanagari, SR; Antony-Debre, I; Mitchell, K; Wang, HR; Kao, YR; Christopeit, M; Carvajal, L; Barreyro, L; Paietta, E; Makishima, H; Will, B; Concha, N; Adams, ND; Schwartz, B; McCabe, MT; Maciejewski, J; Verma, A; Steidl, U New IDH1 mutant inhibitors for treatment of acute myeloid leukemia Nat Chem Biol11:878-86 (2015) [PubMed] Article