| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lysine-specific demethylase 5A |
|---|
| Ligand | BDBM60875 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | KDM5 TR-FRET Assay |
|---|
| pH | 7±n/a |
|---|
| IC50 | 2900±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Vinogradova, M; Gehling, VS; Gustafson, A; Arora, S; Tindell, CA; Wilson, C; Williamson, KE; Guler, GD; Gangurde, P; Manieri, W; Busby, J; Flynn, EM; Lan, F; Kim, HJ; Odate, S; Cochran, AG; Liu, Y; Wongchenko, M; Yang, Y; Cheung, TK; Maile, TM; Lau, T; Costa, M; Hegde, GV; Jackson, E; Pitti, R; Arnott, D; Bailey, C; Bellon, S; Cummings, RT; Albrecht, BK; Harmange, JC; Kiefer, JR; Trojer, P; Classon, M An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat Chem Biol12:531-8 (2016) [PubMed] Article Vinogradova, M; Gehling, VS; Gustafson, A; Arora, S; Tindell, CA; Wilson, C; Williamson, KE; Guler, GD; Gangurde, P; Manieri, W; Busby, J; Flynn, EM; Lan, F; Kim, HJ; Odate, S; Cochran, AG; Liu, Y; Wongchenko, M; Yang, Y; Cheung, TK; Maile, TM; Lau, T; Costa, M; Hegde, GV; Jackson, E; Pitti, R; Arnott, D; Bailey, C; Bellon, S; Cummings, RT; Albrecht, BK; Harmange, JC; Kiefer, JR; Trojer, P; Classon, M An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat Chem Biol12:531-8 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lysine-specific demethylase 5A |
|---|
| Name: | Lysine-specific demethylase 5A |
|---|
| Synonyms: | Histone demethylase JARID1A | JARID1A | Jumonji/ARID domain-containing protein 1A | KDM5A | KDM5A_HUMAN | Lysine-specific demethylase 5A (KDM5A) | RBBP-2 | RBBP2 | RBP2 | Retinoblastoma-binding protein 2 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 192091.79 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P29375 |
|---|
| Residue: | 1690 |
|---|
| Sequence: | MAGVGPGGYAAEFVPPPECPVFEPSWEEFTDPLSFIGRIRPLAEKTGICKIRPPKDWQPP
FACEVKSFRFTPRVQRLNELEAMTRVRLDFLDQLAKFWELQGSTLKIPVVERKILDLYAL
SKIVASKGGFEMVTKEKKWSKVGSRLGYLPGKGTGSLLKSHYERILYPYELFQSGVSLMG
VQMPNLDLKEKVEPEVLSTDTQTSPEPGTRMNILPKRTRRVKTQSESGDVSRNTELKKLQ
IFGAGPKVVGLAMGTKDKEDEVTRRRKVTNRSDAFNMQMRQRKGTLSVNFVDLYVCMFCG
RGNNEDKLLLCDGCDDSYHTFCLIPPLPDVPKGDWRCPKCVAEECSKPREAFGFEQAVRE
YTLQSFGEMADNFKSDYFNMPVHMVPTELVEKEFWRLVSSIEEDVIVEYGADISSKDFGS
GFPVKDGRRKILPEEEEYALSGWNLNNMPVLEQSVLAHINVDISGMKVPWLYVGMCFSSF
CWHIEDHWSYSINYLHWGEPKTWYGVPSHAAEQLEEVMRELAPELFESQPDLLHQLVTIM
NPNVLMEHGVPVYRTNQCAGEFVVTFPRAYHSGFNQGYNFAEAVNFCTADWLPIGRQCVN
HYRRLRRHCVFSHEELIFKMAADPECLDVGLAAMVCKELTLMTEEETRLRESVVQMGVLM
SEEEVFELVPDDERQCSACRTTCFLSALTCSCNPERLVCLYHPTDLCPCPMQKKCLRYRY
PLEDLPSLLYGVKVRAQSYDTWVSRVTEALSANFNHKKDLIELRVMLEDAEDRKYPENDL
FRKLRDAVKEAETCASVAQLLLSKKQKHRQSPDSGRTRTKLTVEELKAFVQQLFSLPCVI
SQARQVKNLLDDVEEFHERAQEAMMDETPDSSKLQMLIDMGSSLYVELPELPRLKQELQQ
ARWLDEVRLTLSDPQQVTLDVMKKLIDSGVGLAPHHAVEKAMAELQELLTVSERWEEKAK
VCLQARPRHSVASLESIVNEAKNIPAFLPNVLSLKEALQKAREWTAKVEAIQSGSNYAYL
EQLESLSAKGRPIPVRLEALPQVESQVAAARAWRERTGRTFLKKNSSHTLLQVLSPRTDI
GVYGSGKNRRKKVKELIEKEKEKDLDLEPLSDLEEGLEETRDTAMVVAVFKEREQKEIEA
MHSLRAANLAKMTMVDRIEEVKFCICRKTASGFMLQCELCKDWFHNSCVPLPKSSSQKKG
SSWQAKEVKFLCPLCMRSRRPRLETILSLLVSLQKLPVRLPEGEALQCLTERAMSWQDRA
RQALATDELSSALAKLSVLSQRMVEQAAREKTEKIISAELQKAAANPDLQGHLPSFQQSA
FNRVVSSVSSSPRQTMDYDDEETDSDEDIRETYGYDMKDTASVKSSSSLEPNLFCDEEIP
IKSEEVVTHMWTAPSFCAEHAYSSASKSCSQGSSTPRKQPRKSPLVPRSLEPPVLELSPG
AKAQLEELMMVGDLLEVSLDETQHIWRILQATHPPSEDRFLHIMEDDSMEEKPLKVKGKD
SSEKKRKRKLEKVEQLFGEGKQKSKELKKMDKPRKKKLKLGADKSKELNKLAKKLAKEEE
RKKKKEKAAAAKVELVKESTEKKREKKVLDIPSKYDWSGAEESDDENAVCAAQNCQRPCK
DKVDWVQCDGGCDEWFHQVCVGVSPEMAENEDYICINCAKKQGPVSPGPAPPPSFIMSYK
LPMEDLKETS
|
|
|
|---|
| BDBM60875 |
|---|
| n/a |
|---|
| Name | BDBM60875 |
|---|
| Synonyms: | 3-((6-(4,5-Dihydro-1H-benzo[d]azepin-3(2H)-yl)-2-(pyridin-2-yl)pyrimidin-4-yl)amino)propanoic acid | 3-{[2-(pyridin-2-yl)-6-(2,3,4,5-tetrahydro-1H-3-benzazepin-3-yl)pyrimidin-4-yl]amino}propanoic acid | GSK J1 | GSK-J1 | GSKJ1 |
|---|
| Type | n/a |
|---|
| Emp. Form. | C22H23N5O2 |
|---|
| Mol. Mass. | 389.4503 |
|---|
| SMILES | OC(=O)CCNc1cc(nc(n1)-c1ccccn1)N1CCc2ccccc2CC1 |
|---|
| Structure | 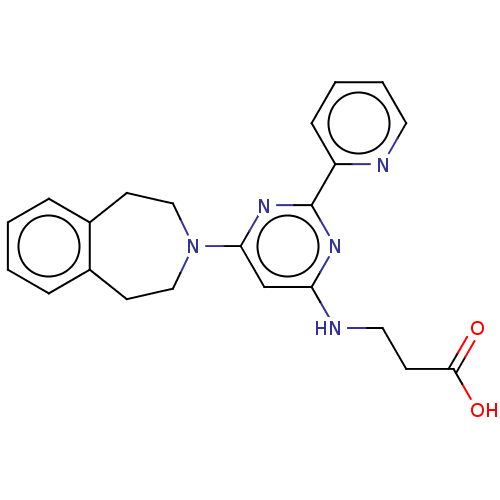
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Vinogradova, M; Gehling, VS; Gustafson, A; Arora, S; Tindell, CA; Wilson, C; Williamson, KE; Guler, GD; Gangurde, P; Manieri, W; Busby, J; Flynn, EM; Lan, F; Kim, HJ; Odate, S; Cochran, AG; Liu, Y; Wongchenko, M; Yang, Y; Cheung, TK; Maile, TM; Lau, T; Costa, M; Hegde, GV; Jackson, E; Pitti, R; Arnott, D; Bailey, C; Bellon, S; Cummings, RT; Albrecht, BK; Harmange, JC; Kiefer, JR; Trojer, P; Classon, M An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat Chem Biol12:531-8 (2016) [PubMed] Article
Vinogradova, M; Gehling, VS; Gustafson, A; Arora, S; Tindell, CA; Wilson, C; Williamson, KE; Guler, GD; Gangurde, P; Manieri, W; Busby, J; Flynn, EM; Lan, F; Kim, HJ; Odate, S; Cochran, AG; Liu, Y; Wongchenko, M; Yang, Y; Cheung, TK; Maile, TM; Lau, T; Costa, M; Hegde, GV; Jackson, E; Pitti, R; Arnott, D; Bailey, C; Bellon, S; Cummings, RT; Albrecht, BK; Harmange, JC; Kiefer, JR; Trojer, P; Classon, M An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat Chem Biol12:531-8 (2016) [PubMed] Article