| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lysine-specific demethylase 2A |
|---|
| Ligand | BDBM26106 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Demethylase AlphaScreen Assay |
|---|
| pH | 7.5±n/a |
|---|
| IC50 | 4.59e+4±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Johansson, C; Velupillai, S; Tumber, A; Szykowska, A; Hookway, ES; Nowak, RP; Strain-Damerell, C; Gileadi, C; Philpott, M; Burgess-Brown, N; Wu, N; Kopec, J; Nuzzi, A; Steuber, H; Egner, U; Badock, V; Munro, S; LaThangue, NB; Westaway, S; Brown, J; Athanasou, N; Prinjha, R; Brennan, PE; Oppermann, U Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol12:539-45 (2016) [PubMed] Article Johansson, C; Velupillai, S; Tumber, A; Szykowska, A; Hookway, ES; Nowak, RP; Strain-Damerell, C; Gileadi, C; Philpott, M; Burgess-Brown, N; Wu, N; Kopec, J; Nuzzi, A; Steuber, H; Egner, U; Badock, V; Munro, S; LaThangue, NB; Westaway, S; Brown, J; Athanasou, N; Prinjha, R; Brennan, PE; Oppermann, U Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol12:539-45 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lysine-specific demethylase 2A |
|---|
| Name: | Lysine-specific demethylase 2A |
|---|
| Synonyms: | AA1-1162 | CXXC8 | F-box and leucine-rich repeat protein 11 | FBL11 | FBL7 | FBXL11 | Homo sapiens lysine demethylase 2A (KDM2A) | JHDM1A | KDM2A | KDM2A_HUMAN | KIAA1004 | Lysine-specific demethylase 2A | Lysine-specific demethylase 2A (KDM2A) | NM_012308 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 132809.04 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 1162 |
|---|
| Sequence: | MEPEEERIRYSQRLRGTMRRRYEDDGISDDEIEGKRTFDLEEKLHTNKYNANFVTFMEGK
DFNVEYIQRGGLRDPLIFKNSDGLGIKMPDPDFTVNDVKMCVGSRRMVDVMDVNTQKGIE
MTMAQWTRYYETPEEEREKLYNVISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKES
QTESTNAILEMQYPKVQKYCLMSVRGCYTDFHVDFGGTSVWYHIHQGGKVFWLIPPTAHN
LELYENWLLSGKQGDIFLGDRVSDCQRIELKQGYTFVIPSGWIHAVYTPTDTLVFGGNFL
HSFNIPMQLKIYNIEDRTRVPNKFRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLS
MDLELNGLESGNGDEEAVDREPRRLSSRRSVLTSPVANGVNLDYDGLGKTCRSLPSLKKT
LAGDSSSDCSRGSHNGQVWDPQCAPRKDRQVHLTHFELEGLRCLVDKLESLPLHKKCVPT
GIEDEDALIADVKILLEELANSDPKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKP
HTMKPAPRLTPVRPAAASPIVSGARRRRVRCRKCKACVQGECGVCHYCRDMKKFGGPGRM
KQSCVLRQCLAPRLPHSVTCSLCGEVDQNEETQDFEKKLMECCICNEIVHPGCLQMDGEG
LLNEELPNCWECPKCYQEDSSEKAQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPT
SMLQLIHDPVSPRGMVTRSSPGAGPSDHHSASRDERFKRRQLLRLQATERTMVREKENNP
SGKKELSEVEKAKIRGSYLTVTLQRPTKELHGTSIVPKLQAITASSANLRHSPRVLVQHC
PARTPQRGDEEGLGGEEEEEEEEEEEDDSAEEGGAARLNGRGSWAQDGDESWMQREVWMS
VFRYLSRRELCECMRVCKTWYKWCCDKRLWTKIDLSRCKAIVPQALSGIIKRQPVSLDLS
WTNISKKQLTWLVNRLPGLKDLLLAGCSWSAVSALSTSSCPLLRTLDLRWAVGIKDPQIR
DLLTPPADKPGQDNRSKLRNMTDFRLAGLDITDATLRLIIRHMPLLSRLDLSHCSHLTDQ
SSNLLTAVGSSTRYSLTELNMAGCNKLTDQTLIYLRRIANVTLIDLRGCKQITRKACEHF
ISDLSINSLYCLSDEKLIQKIS
|
|
|
|---|
| BDBM26106 |
|---|
| n/a |
|---|
| Name | BDBM26106 |
|---|
| Synonyms: | CHEMBL90852 | N-oxalyl glycine, 1a | NOG | Oxalylglycine | [(carboxymethyl)carbamoyl]formic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C4H5NO5 |
|---|
| Mol. Mass. | 147.0862 |
|---|
| SMILES | OC(=O)CNC(=O)C(O)=O |
|---|
| Structure | 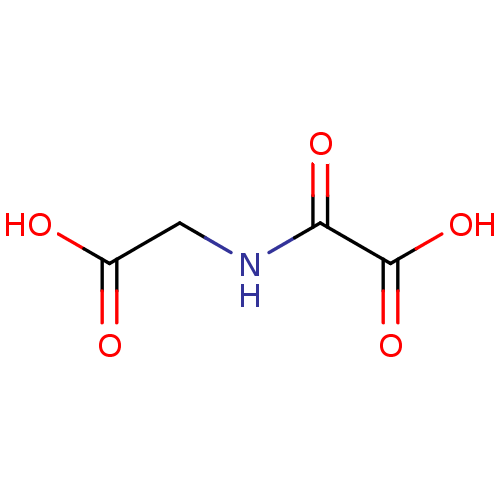
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Johansson, C; Velupillai, S; Tumber, A; Szykowska, A; Hookway, ES; Nowak, RP; Strain-Damerell, C; Gileadi, C; Philpott, M; Burgess-Brown, N; Wu, N; Kopec, J; Nuzzi, A; Steuber, H; Egner, U; Badock, V; Munro, S; LaThangue, NB; Westaway, S; Brown, J; Athanasou, N; Prinjha, R; Brennan, PE; Oppermann, U Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol12:539-45 (2016) [PubMed] Article
Johansson, C; Velupillai, S; Tumber, A; Szykowska, A; Hookway, ES; Nowak, RP; Strain-Damerell, C; Gileadi, C; Philpott, M; Burgess-Brown, N; Wu, N; Kopec, J; Nuzzi, A; Steuber, H; Egner, U; Badock, V; Munro, S; LaThangue, NB; Westaway, S; Brown, J; Athanasou, N; Prinjha, R; Brennan, PE; Oppermann, U Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol12:539-45 (2016) [PubMed] Article