| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Acyl-protein thioesterase 1 [Q83P] |
|---|
| Ligand | BDBM207991 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Steady-State Kinetic Assays |
|---|
| pH | 6.5±n/a |
|---|
| Ki | 370±20 nM |
|---|
| Comments | extracted |
|---|
| Citation |  Won, SJ; Davda, D; Labby, KJ; Hwang, SY; Pricer, R; Majmudar, JD; Armacost, KA; Rodriguez, LA; Rodriguez, CL; Chong, FS; Torossian, KA; Palakurthi, J; Hur, ES; Meagher, JL; Brooks, CL; Stuckey, JA; Martin, BR Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2). ACS Chem Biol11:3374-3382 (2016) [PubMed] Article Won, SJ; Davda, D; Labby, KJ; Hwang, SY; Pricer, R; Majmudar, JD; Armacost, KA; Rodriguez, LA; Rodriguez, CL; Chong, FS; Torossian, KA; Palakurthi, J; Hur, ES; Meagher, JL; Brooks, CL; Stuckey, JA; Martin, BR Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2). ACS Chem Biol11:3374-3382 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Acyl-protein thioesterase 1 [Q83P] |
|---|
| Name: | Acyl-protein thioesterase 1 [Q83P] |
|---|
| Synonyms: | APT1 | Acyl-protein thioesterase 1 (APT1 Q83P) | LPL1 | LYPA1_HUMAN | LYPLA1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 24639.32 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Human APT1 with Q83P mutation |
|---|
| Residue: | 230 |
|---|
| Sequence: | MCGNNMSTPLPAIVPAARKATAAVIFLHGLGDTGHGWAEAFAGIRSSHIKYICPHAPVRP
VTLNMNVAMPSWFDIIGLSPDSPEDESGIKQAAENIKALIDQEVKNGIPSNRIILGGFSQ
GGALSLYTALTTQQKLAGVTALSCWLPLRASFPQGPIGGANRDISILQCHGDCDPLVPLM
FGSLTVEKLKTLVNPANVTFKTYEGMMHSSCQQEMMDVKQFIDKLLPPID
|
|
|
|---|
| BDBM207991 |
|---|
| n/a |
|---|
| Name | BDBM207991 |
|---|
| Synonyms: | ML348 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H17ClF3N3O3 |
|---|
| Mol. Mass. | 415.794 |
|---|
| SMILES | FC(F)(F)c1ccc(Cl)c(NC(=O)CN2CCN(CC2)C(=O)c2ccco2)c1 |
|---|
| Structure | 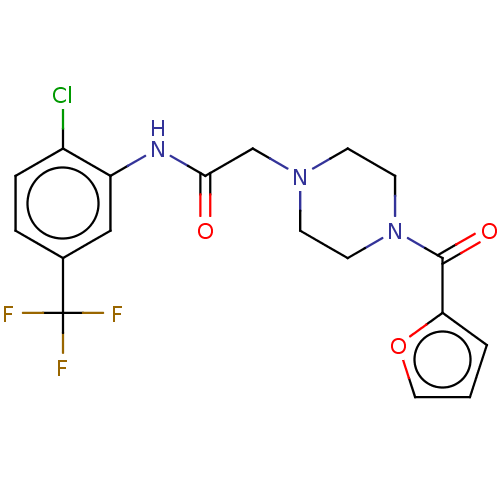
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Won, SJ; Davda, D; Labby, KJ; Hwang, SY; Pricer, R; Majmudar, JD; Armacost, KA; Rodriguez, LA; Rodriguez, CL; Chong, FS; Torossian, KA; Palakurthi, J; Hur, ES; Meagher, JL; Brooks, CL; Stuckey, JA; Martin, BR Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2). ACS Chem Biol11:3374-3382 (2016) [PubMed] Article
Won, SJ; Davda, D; Labby, KJ; Hwang, SY; Pricer, R; Majmudar, JD; Armacost, KA; Rodriguez, LA; Rodriguez, CL; Chong, FS; Torossian, KA; Palakurthi, J; Hur, ES; Meagher, JL; Brooks, CL; Stuckey, JA; Martin, BR Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2). ACS Chem Biol11:3374-3382 (2016) [PubMed] Article