| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Insulin-degrading enzyme |
|---|
| Ligand | BDBM209027 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | IDE Activity Assay |
|---|
| IC50 | 1.5e+3±n/a nM |
|---|
| Citation |  Liu, DR; Maianti, JP; Saghatelian, A; Kleiner, RE Macrocyclic insulin-degrading enzyme (IDE) inhibitors and uses thereof US Patent US9243038 Publication Date 1/26/2016 Liu, DR; Maianti, JP; Saghatelian, A; Kleiner, RE Macrocyclic insulin-degrading enzyme (IDE) inhibitors and uses thereof US Patent US9243038 Publication Date 1/26/2016 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Insulin-degrading enzyme |
|---|
| Name: | Insulin-degrading enzyme |
|---|
| Synonyms: | Abeta-degrading protease | Human Insulin Degrading Enzyme (hIDE) | IDE | IDE_HUMAN | Insulin degrading enzyme (hIDE) | Insulin protease | Insulin-degrading enzyme | Insulinase | Insulysin | insulin-degrading enzyme isoform 1 precursor |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 117968.59 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P14735 |
|---|
| Residue: | 1019 |
|---|
| Sequence: | MRYRLAWLLHPALPSTFRSVLGARLPPPERLCGFQKKTYSKMNNPAIKRIGNHITKSPED
KREYRGLELANGIKVLLISDPTTDKSSAALDVHIGSLSDPPNIAGLSHFCEHMLFLGTKK
YPKENEYSQFLSEHAGSSNAFTSGEHTNYYFDVSHEHLEGALDRFAQFFLCPLFDESCKD
REVNAVDSEHEKNVMNDAWRLFQLEKATGNPKHPFSKFGTGNKYTLETRPNQEGIDVRQE
LLKFHSAYYSSNLMAVCVLGRESLDDLTNLVVKLFSEVENKNVPLPEFPEHPFQEEHLKQ
LYKIVPIKDIRNLYVTFPIPDLQKYYKSNPGHYLGHLIGHEGPGSLLSELKSKGWVNTLV
GGQKEGARGFMFFIINVDLTEEGLLHVEDIILHMFQYIQKLRAEGPQEWVFQECKDLNAV
AFRFKDKERPRGYTSKIAGILHYYPLEEVLTAEYLLEEFRPDLIEMVLDKLRPENVRVAI
VSKSFEGKTDRTEEWYGTQYKQEAIPDEVIKKWQNADLNGKFKLPTKNEFIPTNFEILPL
EKEATPYPALIKDTAMSKLWFKQDDKFFLPKACLNFEFFSPFAYVDPLHCNMAYLYLELL
KDSLNEYAYAAELAGLSYDLQNTIYGMYLSVKGYNDKQPILLKKIIEKMATFEIDEKRFE
IIKEAYMRSLNNFRAEQPHQHAMYYLRLLMTEVAWTKDELKEALDDVTLPRLKAFIPQLL
SRLHIEALLHGNITKQAALGIMQMVEDTLIEHAHTKPLLPSQLVRYREVQLPDRGWFVYQ
QRNEVHNNCGIEIYYQTDMQSTSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRRA
NGIQGLRFIIQSEKPPHYLESRVEAFLITMEKSIEDMTEEAFQKHIQALAIRRLDKPKKL
SAECAKYWGEIISQQYNFDRDNTEVAYLKTLTKEDIIKFYKEMLAVDAPRRHKVSVHVLA
REMDSCPVVGEFPCQNDINLSQAPALPQPEVIQNMTEFKRGLPLFPLVKPHINFMAAKL
|
|
|
|---|
| BDBM209027 |
|---|
| n/a |
|---|
| Name | BDBM209027 |
|---|
| Synonyms: | US9243038, 3b (trans olefin) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C46H62N6O8 |
|---|
| Mol. Mass. | 827.0199 |
|---|
| SMILES | NC(=O)[C@@H]1CCCCNC(=O)[C@@H](Cc2ccc(cc2)C(=O)c2ccccc2)NC(=O)[C@H](CC2CCCCC2)NC(=O)C[C@@H](O)[C@H](CC2CCCCC2)NC(=O)\C=C\C(=O)N1 |r,w:57.60,t:59| |
|---|
| Structure | 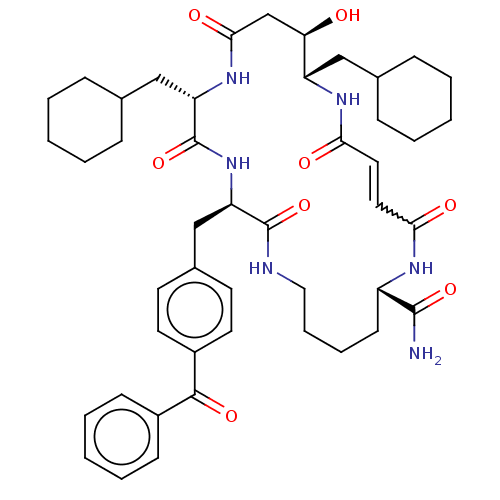
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Liu, DR; Maianti, JP; Saghatelian, A; Kleiner, RE Macrocyclic insulin-degrading enzyme (IDE) inhibitors and uses thereof US Patent US9243038 Publication Date 1/26/2016
Liu, DR; Maianti, JP; Saghatelian, A; Kleiner, RE Macrocyclic insulin-degrading enzyme (IDE) inhibitors and uses thereof US Patent US9243038 Publication Date 1/26/2016