| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Coproheme decarboxylase |
|---|
| Ligand | BDBM209868 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | UV-Visible Titration Assay |
|---|
| pH | 6.8±n/a |
|---|
| Temperature | 298.15±n/a K |
|---|
| Kd | 720±0.21 nM |
|---|
| Comments | extracted |
|---|
| Citation |  Mayfield, JA; Hammer, ND; Kurker, RC; Chen, TK; Ojha, S; Skaar, EP; DuBois, JL The chlorite dismutase (HemQ) from Staphylococcus aureus has a redox-sensitive heme and is associated with the small colony variant phenotype. J Biol Chem288:23488-504 (2013) [PubMed] Article Mayfield, JA; Hammer, ND; Kurker, RC; Chen, TK; Ojha, S; Skaar, EP; DuBois, JL The chlorite dismutase (HemQ) from Staphylococcus aureus has a redox-sensitive heme and is associated with the small colony variant phenotype. J Biol Chem288:23488-504 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Coproheme decarboxylase |
|---|
| Name: | Coproheme decarboxylase |
|---|
| Synonyms: | CHDC_STAAB | Chlorite dismutase (HemQ) | chdC |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 29320.74 |
|---|
| Organism: | Staphylococcus aureus |
|---|
| Description: | Q2YS77 |
|---|
| Residue: | 250 |
|---|
| Sequence: | MSQAAETLDGWYSLHLFYAVDWASLRLAPKDERDALVTELQSFLENTATVRSSKSGDQAI
YNITGQKADLLLWFLRPEMKSLNHIENEFNKLRIADFLIPTYSYVSVIELSNYLAGKSDE
DPYENPHIKARLYPELPHSDYICFYPMNKRRNETYNWYMLTMEERQKLMYDHGMIGRKYA
GKIKQFITGSVGFDDFEWGVTLFSDDVLQFKKIVYEMRFDETTARYGEFGSFFVGHIINT
NEFDQFFAIS
|
|
|
|---|
| BDBM209868 |
|---|
| n/a |
|---|
| Name | BDBM209868 |
|---|
| Synonyms: | 3-[18-(2-carboxyethyl)-8,13-bis(ethenyl)-3,7,12,17-tetramethylporphyrin-21,24-diid-2-yl]propanoic acid;iron(2+) | Haemin | Heme |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C34H32N4O4 |
|---|
| Mol. Mass. | 560.6434 |
|---|
| SMILES | Cc1c(C=C)c2cc3[n-]c(cc4[n-]c(cc5nc(cc1n2)c(C=C)c5C)c(C)c4CCC(O)=O)c(CCC(O)=O)c3C |
|---|
| Structure | 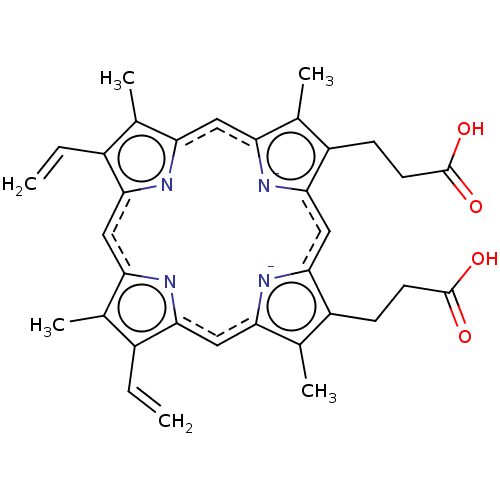
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Mayfield, JA; Hammer, ND; Kurker, RC; Chen, TK; Ojha, S; Skaar, EP; DuBois, JL The chlorite dismutase (HemQ) from Staphylococcus aureus has a redox-sensitive heme and is associated with the small colony variant phenotype. J Biol Chem288:23488-504 (2013) [PubMed] Article
Mayfield, JA; Hammer, ND; Kurker, RC; Chen, TK; Ojha, S; Skaar, EP; DuBois, JL The chlorite dismutase (HemQ) from Staphylococcus aureus has a redox-sensitive heme and is associated with the small colony variant phenotype. J Biol Chem288:23488-504 (2013) [PubMed] Article