| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 6 |
|---|
| Ligand | BDBM19149 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Enzymatic HDAC Activity Assay |
|---|
| pH | 7.5±n/a |
|---|
| IC50 | 9.44±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Lauffer, BE; Mintzer, R; Fong, R; Mukund, S; Tam, C; Zilberleyb, I; Flicke, B; Ritscher, A; Fedorowicz, G; Vallero, R; Ortwine, DF; Gunzner, J; Modrusan, Z; Neumann, L; Koth, CM; Lupardus, PJ; Kaminker, JS; Heise, CE; Steiner, P Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J Biol Chem288:26926-43 (2013) [PubMed] Article Lauffer, BE; Mintzer, R; Fong, R; Mukund, S; Tam, C; Zilberleyb, I; Flicke, B; Ritscher, A; Fedorowicz, G; Vallero, R; Ortwine, DF; Gunzner, J; Modrusan, Z; Neumann, L; Koth, CM; Lupardus, PJ; Kaminker, JS; Heise, CE; Steiner, P Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J Biol Chem288:26926-43 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 6 |
|---|
| Name: | Histone deacetylase 6 |
|---|
| Synonyms: | Cereblon/Histone deacetylase 6 | HD6 | HDAC6 | HDAC6_HUMAN | Histone deacetylase 6 (HDAC6) | Human HDAC6 | KIAA0901 | ORF Names:JM21 |
|---|
| Type: | Chromatin regulator; hydrolase; repressor |
|---|
| Mol. Mass.: | 131381.51 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9UBN7 |
|---|
| Residue: | 1215 |
|---|
| Sequence: | MTSTGQDSTTTRQRRSRQNPQSPPQDSSVTSKRNIKKGAVPRSIPNLAEVKKKGKMKKLG
QAMEEDLIVGLQGMDLNLEAEALAGTGLVLDEQLNEFHCLWDDSFPEGPERLHAIKEQLI
QEGLLDRCVSFQARFAEKEELMLVHSLEYIDLMETTQYMNEGELRVLADTYDSVYLHPNS
YSCACLASGSVLRLVDAVLGAEIRNGMAIIRPPGHHAQHSLMDGYCMFNHVAVAARYAQQ
KHRIRRVLIVDWDVHHGQGTQFTFDQDPSVLYFSIHRYEQGRFWPHLKASNWSTTGFGQG
QGYTINVPWNQVGMRDADYIAAFLHVLLPVALEFQPQLVLVAAGFDALQGDPKGEMAATP
AGFAQLTHLLMGLAGGKLILSLEGGYNLRALAEGVSASLHTLLGDPCPMLESPGAPCRSA
QASVSCALEALEPFWEVLVRSTETVERDNMEEDNVEESEEEGPWEPPVLPILTWPVLQSR
TGLVYDQNMMNHCNLWDSHHPEVPQRILRIMCRLEELGLAGRCLTLTPRPATEAELLTCH
SAEYVGHLRATEKMKTRELHRESSNFDSIYICPSTFACAQLATGAACRLVEAVLSGEVLN
GAAVVRPPGHHAEQDAACGFCFFNSVAVAARHAQTISGHALRILIVDWDVHHGNGTQHMF
EDDPSVLYVSLHRYDHGTFFPMGDEGASSQIGRAAGTGFTVNVAWNGPRMGDADYLAAWH
RLVLPIAYEFNPELVLVSAGFDAARGDPLGGCQVSPEGYAHLTHLLMGLASGRIILILEG
GYNLTSISESMAACTRSLLGDPPPLLTLPRPPLSGALASITETIQVHRRYWRSLRVMKVE
DREGPSSSKLVTKKAPQPAKPRLAERMTTREKKVLEAGMGKVTSASFGEESTPGQTNSET
AVVALTQDQPSEAATGGATLAQTISEAAIGGAMLGQTTSEEAVGGATPDQTTSEETVGGA
ILDQTTSEDAVGGATLGQTTSEEAVGGATLAQTTSEAAMEGATLDQTTSEEAPGGTELIQ
TPLASSTDHQTPPTSPVQGTTPQISPSTLIGSLRTLELGSESQGASESQAPGEENLLGEA
AGGQDMADSMLMQGSRGLTDQAIFYAVTPLPWCPHLVAVCPIPAAGLDVTQPCGDCGTIQ
ENWVCLSCYQVYCGRYINGHMLQHHGNSGHPLVLSYIDLSAWCYYCQAYVHHQALLDVKN
IAHQNKFGEDMPHPH
|
|
|
|---|
| BDBM19149 |
|---|
| n/a |
|---|
| Name | BDBM19149 |
|---|
| Synonyms: | CHEMBL98 | N-hydroxy-N'-phenyloctanediamide | SAHA | US10011611, SAHA | US10188756, Compound SAHA | US11207431, SAHA | US11505523, Compound SAHA | US9115116, SAHA | US9353061, SAHA | US9428447, SAHA | US9695181, Vorinostat | Vorinostat | Zolinza | cid_5311 | suberoylanilide hydroxamic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H20N2O3 |
|---|
| Mol. Mass. | 264.3202 |
|---|
| SMILES | ONC(=O)CCCCCCC(=O)Nc1ccccc1 |
|---|
| Structure | 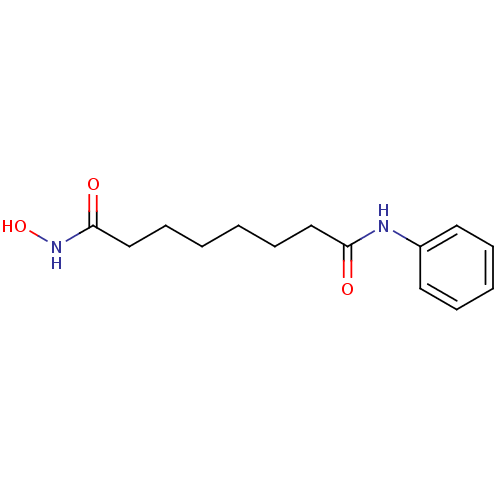
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lauffer, BE; Mintzer, R; Fong, R; Mukund, S; Tam, C; Zilberleyb, I; Flicke, B; Ritscher, A; Fedorowicz, G; Vallero, R; Ortwine, DF; Gunzner, J; Modrusan, Z; Neumann, L; Koth, CM; Lupardus, PJ; Kaminker, JS; Heise, CE; Steiner, P Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J Biol Chem288:26926-43 (2013) [PubMed] Article
Lauffer, BE; Mintzer, R; Fong, R; Mukund, S; Tam, C; Zilberleyb, I; Flicke, B; Ritscher, A; Fedorowicz, G; Vallero, R; Ortwine, DF; Gunzner, J; Modrusan, Z; Neumann, L; Koth, CM; Lupardus, PJ; Kaminker, JS; Heise, CE; Steiner, P Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J Biol Chem288:26926-43 (2013) [PubMed] Article