| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lysine-specific demethylase 5D |
|---|
| Ligand | BDBM223320 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | AlphaScreen Assay |
|---|
| IC50 | 69±0.0 nM |
|---|
| Citation |  Tumber, A; Nuzzi, A; Hookway, ES; Hatch, SB; Velupillai, S; Johansson, C; Kawamura, A; Savitsky, P; Yapp, C; Szykowska, A; Wu, N; Bountra, C; Strain-Damerell, C; Burgess-Brown, NA; Ruda, GF; Fedorov, O; Munro, S; England, KS; Nowak, RP; Schofield, CJ; La Thangue, NB; Pawlyn, C; Davies, F; Morgan, G; Athanasou, N; Müller, S; Oppermann, U; Brennan, PE Potent and Selective KDM5 Inhibitor Stops Cellular Demethylation of H3K4me3 at Transcription Start Sites and Proliferation of MM1S Myeloma Cells. Cell Chem Biol24:371-380 (2017) [PubMed] Article Tumber, A; Nuzzi, A; Hookway, ES; Hatch, SB; Velupillai, S; Johansson, C; Kawamura, A; Savitsky, P; Yapp, C; Szykowska, A; Wu, N; Bountra, C; Strain-Damerell, C; Burgess-Brown, NA; Ruda, GF; Fedorov, O; Munro, S; England, KS; Nowak, RP; Schofield, CJ; La Thangue, NB; Pawlyn, C; Davies, F; Morgan, G; Athanasou, N; Müller, S; Oppermann, U; Brennan, PE Potent and Selective KDM5 Inhibitor Stops Cellular Demethylation of H3K4me3 at Transcription Start Sites and Proliferation of MM1S Myeloma Cells. Cell Chem Biol24:371-380 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lysine-specific demethylase 5D |
|---|
| Name: | Lysine-specific demethylase 5D |
|---|
| Synonyms: | HY | HYA | JARID1D | KDM5D | KDM5D_HUMAN | KIAA0234 | Lysine-specific demethylase 5D (KDM5D) | SMCY |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 174050.91 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9BY66 |
|---|
| Residue: | 1539 |
|---|
| Sequence: | MEPGCDEFLPPPECPVFEPSWAEFQDPLGYIAKIRPIAEKSGICKIRPPADWQPPFAVEV
DNFRFTPRVQRLNELEAQTRVKLNYLDQIAKFWEIQGSSLKIPNVERKILDLYSLSKIVI
EEGGYEAICKDRRWARVAQRLHYPPGKNIGSLLRSHYERIIYPYEMFQSGANHVQCNTHP
FDNEVKDKEYKPHSIPLRQSVQPSKFSSYSRRAKRLQPDPEPTEEDIEKHPELKKLQIYG
PGPKMMGLGLMAKDKDKTVHKKVTCPPTVTVKDEQSGGGNVSSTLLKQHLSLEPCTKTTM
QLRKNHSSAQFIDSYICQVCSRGDEDDKLLFCDGCDDNYHIFCLLPPLPEIPRGIWRCPK
CILAECKQPPEAFGFEQATQEYSLQSFGEMADSFKSDYFNMPVHMVPTELVEKEFWRLVS
SIEEDVTVEYGADIHSKEFGSGFPVSNSKQNLSPEEKEYATSGWNLNVMPVLDQSVLCHI
NADISGMKVPWLYVGMVFSAFCWHIEDHWSYSINYLHWGEPKTWYGVPSLAAEHLEEVMK
MLTPELFDSQPDLLHQLVTLMNPNTLMSHGVPVVRTNQCAGEFVITFPRAYHSGFNQGYN
FAEAVNFCTADWLPAGRQCIEHYRRLRRYCVFSHEELICKMAAFPETLDLNLAVAVHKEM
FIMVQEERRLRKALLEKGVTEAEREAFELLPDDERQCIKCKTTCFLSALACYDCPDGLVC
LSHINDLCKCSSSRQYLRYRYTLDELPTMLHKLKIRAESFDTWANKVRVALEVEDGRKRS
FEELRALESEARERRFPNSELLQRLKNCLSEVEACIAQVLGLVSGQVARMDTPQLTLTEL
RVLLEQMGSLPCAMHQIGDVKDVLEQVEAYQAEAREALATLPSSPGLLRSLLERGQQLGV
EVPEAHQLQQQVEQAQWLDEVKQALAPSAHRGSLVIMQGLLVMGAKIASSPSVDKARAEL
QELLTIAERWEEKAHFCLEARQKHPPATLEAIIRETENIPVHLPNIQALKEALTKAQAWI
ADVDEIQNGDHYPCLDDLEGLVAVGRDLPVGLEELRQLELQVLTAHSWREKASKTFLKKN
SCYTLLEVLCPCADAGSDSTKRSRWMEKALGLYQCDTELLGLSAQDLRDPGSVIVAFKEG
EQKEKEGILQLRRTNSAKPSPLAPSLMASSPTSICVCGQVPAGVGVLQCDLCQDWFHGQC
VSVPHLLTSPKPSLTSSPLLAWWEWDTKFLCPLCMRSRRPRLETILALLVALQRLPVRLP
EGEALQCLTERAIGWQDRARKALASEDVTALLRQLAELRQQLQAKPRPEEASVYTSATAC
DPIREGSGNNISKVQGLLENGDSVTSPENMAPGKGSDLELLSSLLPQLTGPVLELPEAIR
APLEELMMEGDLLEVTLDENHSIWQLLQAGQPPDLDRIRTLLELEKFEHQGSRTRSRALE
RRRRRQKVDQGRNVENLVQQELQSKRARSSGIMSQVGREEEHYQEKADRENMFLTPSTDH
SPFLKGNQNSLQHKDSGSSAACPSLMPLLQLSYSDEQQL
|
|
|
|---|
| BDBM223320 |
|---|
| n/a |
|---|
| Name | BDBM223320 |
|---|
| Synonyms: | KDOAM-25 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H25N5O2 |
|---|
| Mol. Mass. | 307.3913 |
|---|
| SMILES | CCN(CCN(C)C)C(=O)CNCc1cc(ccn1)C(N)=O |
|---|
| Structure | 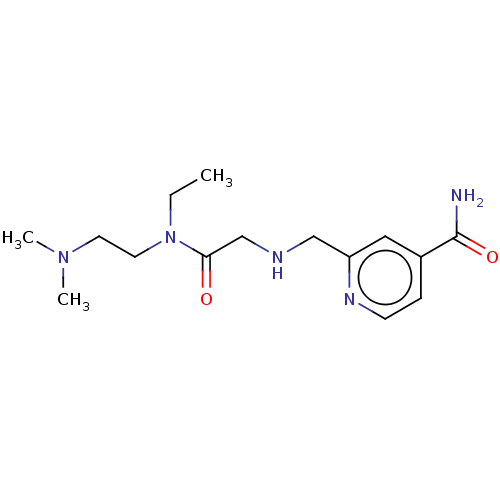
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Tumber, A; Nuzzi, A; Hookway, ES; Hatch, SB; Velupillai, S; Johansson, C; Kawamura, A; Savitsky, P; Yapp, C; Szykowska, A; Wu, N; Bountra, C; Strain-Damerell, C; Burgess-Brown, NA; Ruda, GF; Fedorov, O; Munro, S; England, KS; Nowak, RP; Schofield, CJ; La Thangue, NB; Pawlyn, C; Davies, F; Morgan, G; Athanasou, N; Müller, S; Oppermann, U; Brennan, PE Potent and Selective KDM5 Inhibitor Stops Cellular Demethylation of H3K4me3 at Transcription Start Sites and Proliferation of MM1S Myeloma Cells. Cell Chem Biol24:371-380 (2017) [PubMed] Article
Tumber, A; Nuzzi, A; Hookway, ES; Hatch, SB; Velupillai, S; Johansson, C; Kawamura, A; Savitsky, P; Yapp, C; Szykowska, A; Wu, N; Bountra, C; Strain-Damerell, C; Burgess-Brown, NA; Ruda, GF; Fedorov, O; Munro, S; England, KS; Nowak, RP; Schofield, CJ; La Thangue, NB; Pawlyn, C; Davies, F; Morgan, G; Athanasou, N; Müller, S; Oppermann, U; Brennan, PE Potent and Selective KDM5 Inhibitor Stops Cellular Demethylation of H3K4me3 at Transcription Start Sites and Proliferation of MM1S Myeloma Cells. Cell Chem Biol24:371-380 (2017) [PubMed] Article