| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mas-related G-protein coupled receptor member X2 |
|---|
| Ligand | BDBM50001450 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Intracellular Calcium Mobilization Assay |
|---|
| pH | 7.4±0 |
|---|
| Temperature | 303.15±0 K |
|---|
| EC50 | 1.1e+3±4.1e+2 nM |
|---|
| Citation |  Lansu, K; Karpiak, J; Liu, J; Huang, XP; McCorvy, JD; Kroeze, WK; Che, T; Nagase, H; Carroll, FI; Jin, J; Shoichet, BK; Roth, BL In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat Chem Biol13:529-536 (2017) [PubMed] Article Lansu, K; Karpiak, J; Liu, J; Huang, XP; McCorvy, JD; Kroeze, WK; Che, T; Nagase, H; Carroll, FI; Jin, J; Shoichet, BK; Roth, BL In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat Chem Biol13:529-536 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mas-related G-protein coupled receptor member X2 |
|---|
| Name: | Mas-related G-protein coupled receptor member X2 |
|---|
| Synonyms: | MRGPRX2 | MRGX2 | MRGX2_HUMAN | Mas-related G protein-coupled receptor X2 (MRGPRX2) | Mas-related G-protein coupled receptor member X2 |
|---|
| Type: | GPCR |
|---|
| Mol. Mass.: | 37104.79 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q96LB1 |
|---|
| Residue: | 330 |
|---|
| Sequence: | MDPTTPAWGTESTTVNGNDQALLLLCGKETLIPVFLILFIALVGLVGNGFVLWLLGFRMR
RNAFSVYVLSLAGADFLFLCFQIINCLVYLSNFFCSISINFPSFFTTVMTCAYLAGLSML
STVSTERCLSVLWPIWYRCRRPRHLSAVVCVLLWALSLLLSILEGKFCGFLFSDGDSGWC
QTFDFITAAWLIFLFMVLCGSSLALLVRILCGSRGLPLTRLYLTILLTVLVFLLCGLPFG
IQWFLILWIWKDSDVLFCHIHPVSVVLSSLNSSANPIIYFFVGSFRKQWRLQQPILKLAL
QRALQDIAEVDHSEGCFRQGTPEMSRSSLV
|
|
|
|---|
| BDBM50001450 |
|---|
| n/a |
|---|
| Name | BDBM50001450 |
|---|
| Synonyms: | (SP)Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2 | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met NH2 | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2 | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2(Substance P) | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2.(Substance P) | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-amine | ArgProLysProGlnGlnPhePheGlyLeuMet | CHEMBL235363 | H-Arg-Pro-Lys-Pro-Gln-Gln-Phe-D-Phe-Gly-Leu-Met-NH2 | H-Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2(Substance P) | Substance P | Substance P (Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-MetNH2) | Substance P analogue | tachykinin substance P (SP) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C63H98N18O13S |
|---|
| Mol. Mass. | 1347.63 |
|---|
| SMILES | [#6]-[#16]-[#6]-[#6]-[#6@H](-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6@H](-[#6]-c1ccccc1)-[#7]-[#6](=O)-[#6@H](-[#6]-c1ccccc1)-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#7]-[#6](=O)-[#6@@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6@@H](-[#7])-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](-[#7])=O |r| |
|---|
| Structure | 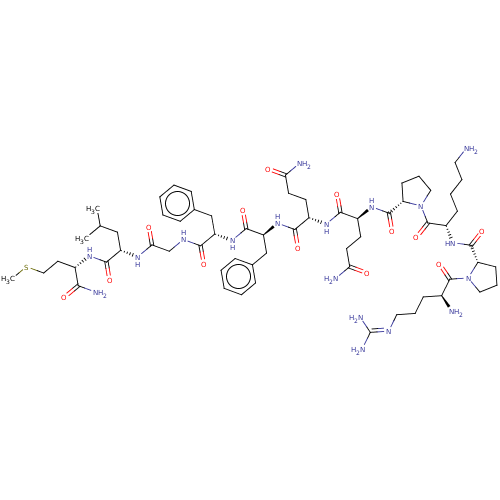
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lansu, K; Karpiak, J; Liu, J; Huang, XP; McCorvy, JD; Kroeze, WK; Che, T; Nagase, H; Carroll, FI; Jin, J; Shoichet, BK; Roth, BL In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat Chem Biol13:529-536 (2017) [PubMed] Article
Lansu, K; Karpiak, J; Liu, J; Huang, XP; McCorvy, JD; Kroeze, WK; Che, T; Nagase, H; Carroll, FI; Jin, J; Shoichet, BK; Roth, BL In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat Chem Biol13:529-536 (2017) [PubMed] Article