| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Heme oxygenase HutZ [D132N] |
|---|
| Ligand | BDBM231681 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Heme Binding Assay |
|---|
| pH | 8±0 |
|---|
| Kd | 1.5e+2± 2e+1 nM |
|---|
| Citation |  Uchida, T; Dojun, N; Sekine, Y; Ishimori, K Heme Proximal Hydrogen Bonding between His170 and Asp132 Plays an Essential Role in the Heme Degradation Reaction of HutZ from Vibrio cholerae. Biochemistry56:2723-2734 (2017) [PubMed] Article Uchida, T; Dojun, N; Sekine, Y; Ishimori, K Heme Proximal Hydrogen Bonding between His170 and Asp132 Plays an Essential Role in the Heme Degradation Reaction of HutZ from Vibrio cholerae. Biochemistry56:2723-2734 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Heme oxygenase HutZ [D132N] |
|---|
| Name: | Heme oxygenase HutZ [D132N] |
|---|
| Synonyms: | HUTZ_VIBCH | Heme-binding protein HutZ (HutZ)(D132N) | hutZ |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 20294.78 |
|---|
| Organism: | Vibrio cholerae |
|---|
| Description: | V. cholerae HutZ variant D132N |
|---|
| Residue: | 176 |
|---|
| Sequence: | MDQQVKQERLQGRLEPEIKEFRQERKTLQLATVDAQGRPNVSYAPFVQNQEGYFVLISHI
ARHARNLEVNPQVSIMMIEDETEAKQLFARKRLTFDAVASMVERDSELWCQVIAQMGERF
GEIIDGLSQLQNFMLFRLQPEQGLFVKGFGQAYQVSGDDLVDFVHLEEGHRKISNG
|
|
|
|---|
| BDBM231681 |
|---|
| n/a |
|---|
| Name | BDBM231681 |
|---|
| Synonyms: | Hemin |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C34H32N4O4 |
|---|
| Mol. Mass. | 560.6434 |
|---|
| SMILES | Cc1c(CCC(O)=O)c2cc3[n-]c(cc4nc(cc5[n-]c(cc1n2)c(C)c5C=C)c(C)c4C=C)c(C)c3CCC(O)=O |
|---|
| Structure | 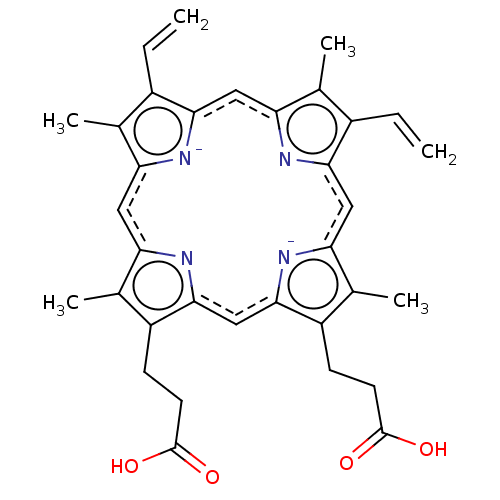
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Uchida, T; Dojun, N; Sekine, Y; Ishimori, K Heme Proximal Hydrogen Bonding between His170 and Asp132 Plays an Essential Role in the Heme Degradation Reaction of HutZ from Vibrio cholerae. Biochemistry56:2723-2734 (2017) [PubMed] Article
Uchida, T; Dojun, N; Sekine, Y; Ishimori, K Heme Proximal Hydrogen Bonding between His170 and Asp132 Plays an Essential Role in the Heme Degradation Reaction of HutZ from Vibrio cholerae. Biochemistry56:2723-2734 (2017) [PubMed] Article