 Found 236 hits of affinity data for UniProtKB/TrEMBL: P02829
Found 236 hits of affinity data for UniProtKB/TrEMBL: P02829 Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM15361
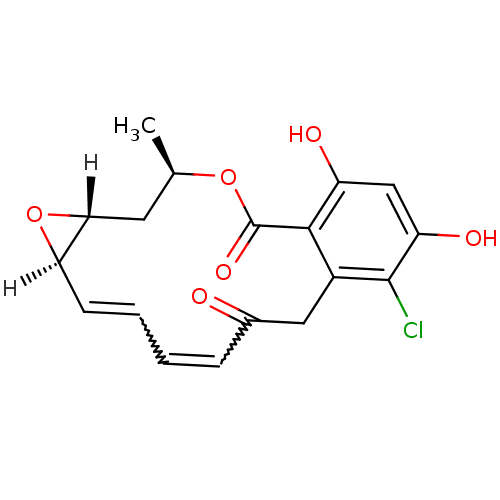
((1aR,2Z,4E,14R,15aR)-8-chloro-9,11-dihydroxy-14-me...)Show SMILES [H][C@@]12C[C@@H](C)OC(=O)c3c(O)cc(O)c(Cl)c3CC(=O)C=CC=C[C@@]1([H])O2 |r,w:22.22,20.20| Show InChI InChI=1S/C18H17ClO6/c1-9-6-15-14(25-15)5-3-2-4-10(20)7-11-16(18(23)24-9)12(21)8-13(22)17(11)19/h2-5,8-9,14-15,21-22H,6-7H2,1H3/t9-,14-,15-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 30 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM15361

((1aR,2Z,4E,14R,15aR)-8-chloro-9,11-dihydroxy-14-me...)Show SMILES [H][C@@]12C[C@@H](C)OC(=O)c3c(O)cc(O)c(Cl)c3CC(=O)C=CC=C[C@@]1([H])O2 |r,w:22.22,20.20| Show InChI InChI=1S/C18H17ClO6/c1-9-6-15-14(25-15)5-3-2-4-10(20)7-11-16(18(23)24-9)12(21)8-13(22)17(11)19/h2-5,8-9,14-15,21-22H,6-7H2,1H3/t9-,14-,15-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 47 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68263

(Microlactone, 15h)Show SMILES Oc1cc(O)c2c(CC(=O)CCCCCC=CCCOC2=O)c1Cl |w:17.16,15.14| Show InChI InChI=1S/C18H21ClO5/c19-17-13-10-12(20)8-6-4-2-1-3-5-7-9-24-18(23)16(13)14(21)11-15(17)22/h3,5,11,21-22H,1-2,4,6-10H2 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 100 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68256
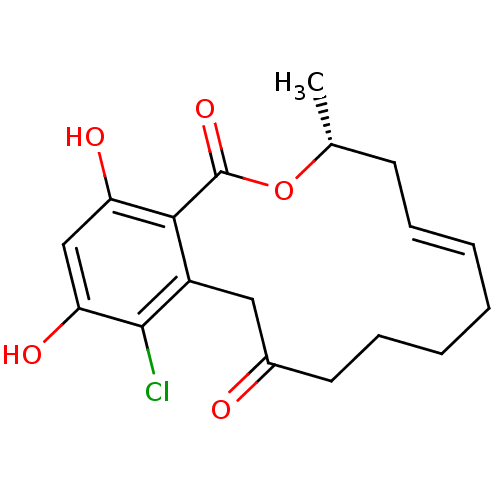
(Microlactone, 15c)Show SMILES C[C@@H]1C\C=C\CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |r,t:3| Show InChI InChI=1S/C18H21ClO5/c1-11-7-5-3-2-4-6-8-12(20)9-13-16(18(23)24-11)14(21)10-15(22)17(13)19/h3,5,10-11,21-22H,2,4,6-9H2,1H3/b5-3+/t11-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | 100 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68248
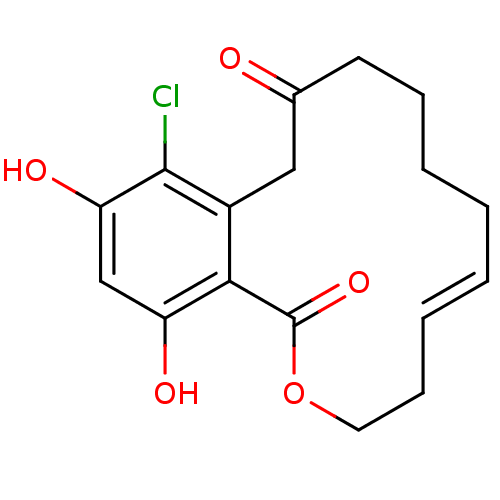
(Microlactone, 15a)Show SMILES Oc1cc(O)c2c(CC(=O)CCCC\C=C\CCOC2=O)c1Cl |t:14| Show InChI InChI=1S/C17H19ClO5/c18-16-12-9-11(19)7-5-3-1-2-4-6-8-23-17(22)15(12)13(20)10-14(16)21/h2,4,10,20-21H,1,3,5-9H2/b4-2+ | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| DrugBank
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | 100 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68248

(Microlactone, 15a)Show SMILES Oc1cc(O)c2c(CC(=O)CCCC\C=C\CCOC2=O)c1Cl |t:14| Show InChI InChI=1S/C17H19ClO5/c18-16-12-9-11(19)7-5-3-1-2-4-6-8-23-17(22)15(12)13(20)10-14(16)21/h2,4,10,20-21H,1,3,5-9H2/b4-2+ | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| DrugBank
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | 120 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68264

(Microlactone, 15i)Show SMILES C[C@@H]1CC=CCCCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |r,w:2.2,4.4| Show InChI InChI=1S/C19H23ClO5/c1-12-8-6-4-2-3-5-7-9-13(21)10-14-17(19(24)25-12)15(22)11-16(23)18(14)20/h4,6,11-12,22-23H,2-3,5,7-10H2,1H3/t12-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 160 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68263

(Microlactone, 15h)Show SMILES Oc1cc(O)c2c(CC(=O)CCCCCC=CCCOC2=O)c1Cl |w:17.16,15.14| Show InChI InChI=1S/C18H21ClO5/c19-17-13-10-12(20)8-6-4-2-1-3-5-7-9-24-18(23)16(13)14(21)11-15(17)22/h3,5,11,21-22H,1-2,4,6-10H2 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 160 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68250
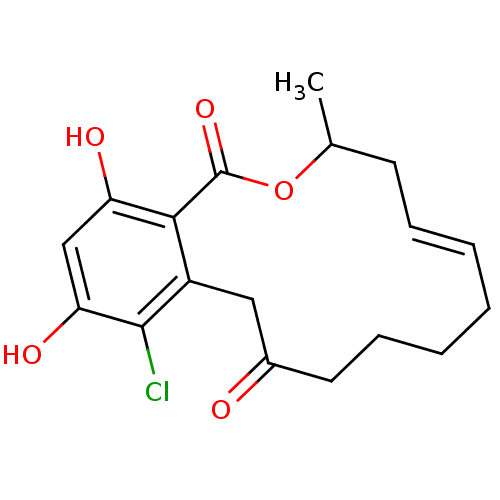
(Microlactone, 15b)Show SMILES CC1C\C=C\CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |t:3| Show InChI InChI=1S/C18H21ClO5/c1-11-7-5-3-2-4-6-8-12(20)9-13-16(18(23)24-11)14(21)10-15(22)17(13)19/h3,5,10-11,21-22H,2,4,6-9H2,1H3/b5-3+ | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | 160 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68249
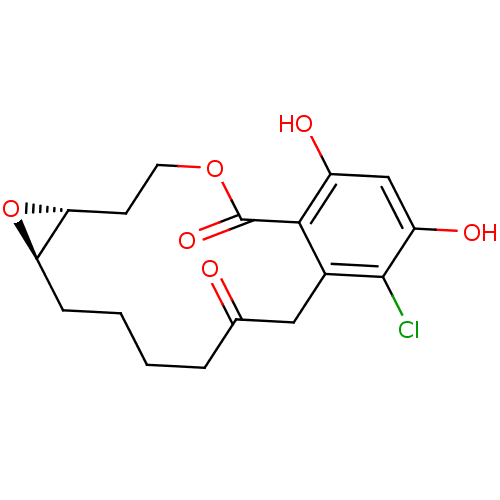
(Microlactone, 16a)Show SMILES Oc1cc(O)c2c(CC(=O)CCCC[C@H]3O[C@@H]3CCOC2=O)c1Cl |r| Show InChI InChI=1S/C17H19ClO6/c18-16-10-7-9(19)3-1-2-4-13-14(24-13)5-6-23-17(22)15(10)11(20)8-12(16)21/h8,13-14,20-21H,1-7H2/t13-,14-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 190 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM15361

((1aR,2Z,4E,14R,15aR)-8-chloro-9,11-dihydroxy-14-me...)Show SMILES [H][C@@]12C[C@@H](C)OC(=O)c3c(O)cc(O)c(Cl)c3CC(=O)C=CC=C[C@@]1([H])O2 |r,w:22.22,20.20| Show InChI InChI=1S/C18H17ClO6/c1-9-6-15-14(25-15)5-3-2-4-10(20)7-11-16(18(23)24-9)12(21)8-13(22)17(11)19/h2-5,8-9,14-15,21-22H,6-7H2,1H3/t9-,14-,15-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 200 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM15361

((1aR,2Z,4E,14R,15aR)-8-chloro-9,11-dihydroxy-14-me...)Show SMILES [H][C@@]12C[C@@H](C)OC(=O)c3c(O)cc(O)c(Cl)c3CC(=O)C=CC=C[C@@]1([H])O2 |r,w:22.22,20.20| Show InChI InChI=1S/C18H17ClO6/c1-9-6-15-14(25-15)5-3-2-4-10(20)7-11-16(18(23)24-9)12(21)8-13(22)17(11)19/h2-5,8-9,14-15,21-22H,6-7H2,1H3/t9-,14-,15-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | <200 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd
| Assay Description
The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... |
Chem Biol 11: 775-85 (2004)
Article DOI: 10.1016/j.chembiol.2004.03.033
BindingDB Entry DOI: 10.7270/Q2RX99BC |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68264

(Microlactone, 15i)Show SMILES C[C@@H]1CC=CCCCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |r,w:2.2,4.4| Show InChI InChI=1S/C19H23ClO5/c1-12-8-6-4-2-3-5-7-9-13(21)10-14-17(19(24)25-12)15(22)11-16(23)18(14)20/h4,6,11-12,22-23H,2-3,5,7-10H2,1H3/t12-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 220 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68266

(Microlactone, 15j)Show SMILES Oc1cc(O)c2c(CC(=O)CCCCCCC=CCCOC2=O)c1Cl |w:18.17,16.15| Show InChI InChI=1S/C19H23ClO5/c20-18-14-11-13(21)9-7-5-3-1-2-4-6-8-10-25-19(24)17(14)15(22)12-16(18)23/h4,6,12,22-23H,1-3,5,7-11H2 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 240 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68251
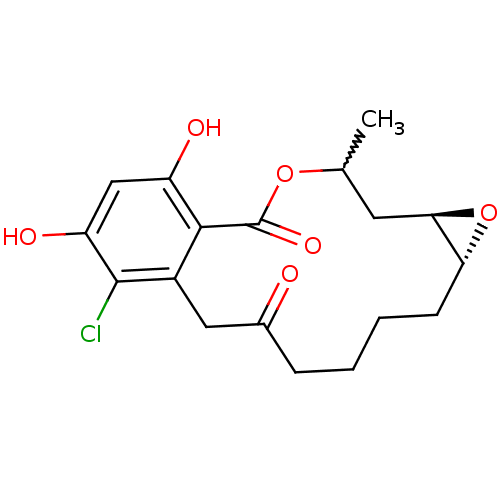
(Microlactone, 16b)Show SMILES CC1C[C@H]2O[C@@H]2CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |r,w:1.0| Show InChI InChI=1S/C18H21ClO6/c1-9-6-15-14(25-15)5-3-2-4-10(20)7-11-16(18(23)24-9)12(21)8-13(22)17(11)19/h8-9,14-15,21-22H,2-7H2,1H3/t9?,14-,15-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 250 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68250

(Microlactone, 15b)Show SMILES CC1C\C=C\CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |t:3| Show InChI InChI=1S/C18H21ClO5/c1-11-7-5-3-2-4-6-8-12(20)9-13-16(18(23)24-11)14(21)10-15(22)17(13)19/h3,5,10-11,21-22H,2,4,6-9H2,1H3/b5-3+ | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | 310 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68249

(Microlactone, 16a)Show SMILES Oc1cc(O)c2c(CC(=O)CCCC[C@H]3O[C@@H]3CCOC2=O)c1Cl |r| Show InChI InChI=1S/C17H19ClO6/c18-16-10-7-9(19)3-1-2-4-13-14(24-13)5-6-23-17(22)15(10)11(20)8-12(16)21/h8,13-14,20-21H,1-7H2/t13-,14-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 310 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68251

(Microlactone, 16b)Show SMILES CC1C[C@H]2O[C@@H]2CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |r,w:1.0| Show InChI InChI=1S/C18H21ClO6/c1-9-6-15-14(25-15)5-3-2-4-10(20)7-11-16(18(23)24-9)12(21)8-13(22)17(11)19/h8-9,14-15,21-22H,2-7H2,1H3/t9?,14-,15-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 310 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68260
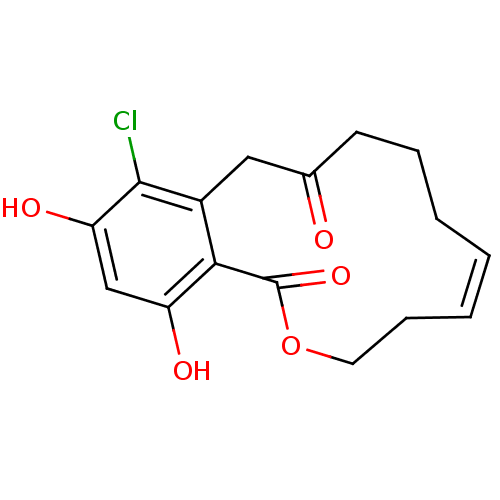
(Microlactone, 15f cis)Show SMILES Oc1cc(O)c2c(CC(=O)CCC\C=C/CCOC2=O)c1Cl |c:13| Show InChI InChI=1S/C16H17ClO5/c17-15-11-8-10(18)6-4-2-1-3-5-7-22-16(21)14(11)12(19)9-13(15)20/h1,3,9,19-20H,2,4-8H2/b3-1- | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| DrugBank
MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 500 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68255
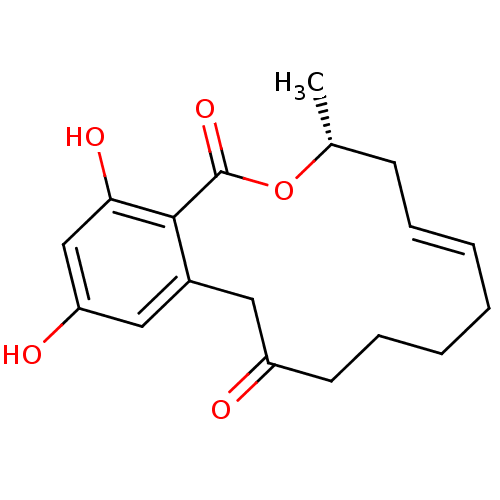
(Microlactone, 14c)Show SMILES C[C@@H]1C\C=C\CCCCC(=O)Cc2cc(O)cc(O)c2C(=O)O1 |r,t:3| Show InChI InChI=1S/C18H22O5/c1-12-7-5-3-2-4-6-8-14(19)9-13-10-15(20)11-16(21)17(13)18(22)23-12/h3,5,10-12,20-21H,2,4,6-9H2,1H3/b5-3+/t12-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 500 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68255

(Microlactone, 14c)Show SMILES C[C@@H]1C\C=C\CCCCC(=O)Cc2cc(O)cc(O)c2C(=O)O1 |r,t:3| Show InChI InChI=1S/C18H22O5/c1-12-7-5-3-2-4-6-8-14(19)9-13-10-15(20)11-16(21)17(13)18(22)23-12/h3,5,10-12,20-21H,2,4,6-9H2,1H3/b5-3+/t12-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 540 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68261

(Microlactone, 15f trans)Show SMILES Oc1cc(O)c2c(CC(=O)CCC\C=C\CCOC2=O)c1Cl |t:13| Show InChI InChI=1S/C16H17ClO5/c17-15-11-8-10(18)6-4-2-1-3-5-7-22-16(21)14(11)12(19)9-13(15)20/h1,3,9,19-20H,2,4-8H2/b3-1+ | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| DrugBank
MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 590 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68263

(Microlactone, 15h)Show SMILES Oc1cc(O)c2c(CC(=O)CCCCCC=CCCOC2=O)c1Cl |w:17.16,15.14| Show InChI InChI=1S/C18H21ClO5/c19-17-13-10-12(20)8-6-4-2-1-3-5-7-9-24-18(23)16(13)14(21)11-15(17)22/h3,5,11,21-22H,1-2,4,6-10H2 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 760 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68260

(Microlactone, 15f cis)Show SMILES Oc1cc(O)c2c(CC(=O)CCC\C=C/CCOC2=O)c1Cl |c:13| Show InChI InChI=1S/C16H17ClO5/c17-15-11-8-10(18)6-4-2-1-3-5-7-22-16(21)14(11)12(19)9-13(15)20/h1,3,9,19-20H,2,4-8H2/b3-1- | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| DrugBank
MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 850 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68266

(Microlactone, 15j)Show SMILES Oc1cc(O)c2c(CC(=O)CCCCCCC=CCCOC2=O)c1Cl |w:18.17,16.15| Show InChI InChI=1S/C19H23ClO5/c20-18-14-11-13(21)9-7-5-3-1-2-4-6-8-10-25-19(24)17(14)15(22)12-16(18)23/h4,6,12,22-23H,1-3,5,7-11H2 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 870 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68250

(Microlactone, 15b)Show SMILES CC1C\C=C\CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |t:3| Show InChI InChI=1S/C18H21ClO5/c1-11-7-5-3-2-4-6-8-12(20)9-13-16(18(23)24-11)14(21)10-15(22)17(13)19/h3,5,10-11,21-22H,2,4,6-9H2,1H3/b5-3+ | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | 1.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68261

(Microlactone, 15f trans)Show SMILES Oc1cc(O)c2c(CC(=O)CCC\C=C\CCOC2=O)c1Cl |t:13| Show InChI InChI=1S/C16H17ClO5/c17-15-11-8-10(18)6-4-2-1-3-5-7-22-16(21)14(11)12(19)9-13(15)20/h1,3,9,19-20H,2,4-8H2/b3-1+ | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| DrugBank
MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 1.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68256

(Microlactone, 15c)Show SMILES C[C@@H]1C\C=C\CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |r,t:3| Show InChI InChI=1S/C18H21ClO5/c1-11-7-5-3-2-4-6-8-12(20)9-13-16(18(23)24-11)14(21)10-15(22)17(13)19/h3,5,10-11,21-22H,2,4,6-9H2,1H3/b5-3+/t11-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | 1.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68256

(Microlactone, 15c)Show SMILES C[C@@H]1C\C=C\CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |r,t:3| Show InChI InChI=1S/C18H21ClO5/c1-11-7-5-3-2-4-6-8-12(20)9-13-16(18(23)24-11)14(21)10-15(22)17(13)19/h3,5,10-11,21-22H,2,4,6-9H2,1H3/b5-3+/t11-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | <1.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68248

(Microlactone, 15a)Show SMILES Oc1cc(O)c2c(CC(=O)CCCC\C=C\CCOC2=O)c1Cl |t:14| Show InChI InChI=1S/C17H19ClO5/c18-16-12-9-11(19)7-5-3-1-2-4-6-8-23-17(22)15(12)13(20)10-14(16)21/h2,4,10,20-21H,1,3,5-9H2/b4-2+ | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| DrugBank
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | <1.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68265
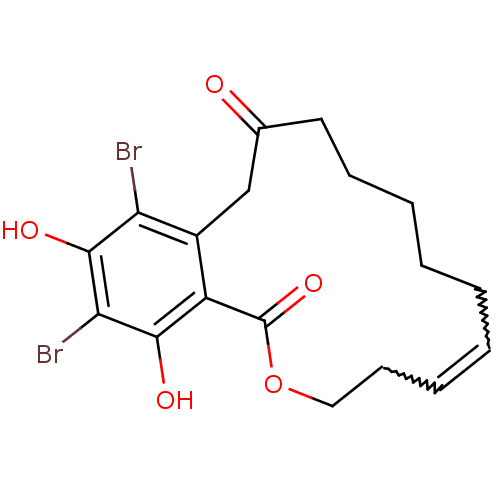
(Microlactone, 18)Show SMILES Oc1c(Br)c(O)c2c(CC(=O)CCCCCC=CCCOC2=O)c1Br |w:18.17,16.15| Show InChI InChI=1S/C18H20Br2O5/c19-14-12-10-11(21)8-6-4-2-1-3-5-7-9-25-18(24)13(12)16(22)15(20)17(14)23/h3,5,22-23H,1-2,4,6-10H2 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 1.10E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68252

(Microlactone, 17)Show InChI InChI=1S/C18H23ClO5/c1-11-7-5-3-2-4-6-8-12(20)9-13-16(18(23)24-11)14(21)10-15(22)17(13)19/h10-11,21-22H,2-9H2,1H3 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 1.29E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68248

(Microlactone, 15a)Show SMILES Oc1cc(O)c2c(CC(=O)CCCC\C=C\CCOC2=O)c1Cl |t:14| Show InChI InChI=1S/C17H19ClO5/c18-16-12-9-11(19)7-5-3-1-2-4-6-8-23-17(22)15(12)13(20)10-14(16)21/h2,4,10,20-21H,1,3,5-9H2/b4-2+ | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| DrugBank
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | 1.30E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68250

(Microlactone, 15b)Show SMILES CC1C\C=C\CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |t:3| Show InChI InChI=1S/C18H21ClO5/c1-11-7-5-3-2-4-6-8-12(20)9-13-16(18(23)24-11)14(21)10-15(22)17(13)19/h3,5,10-11,21-22H,2,4,6-9H2,1H3/b5-3+ | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | 1.50E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM50409989
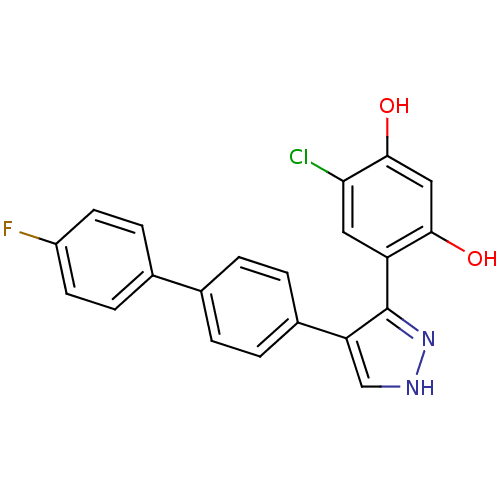
(CHEMBL361941)Show SMILES Oc1cc(O)c(cc1Cl)-c1n[nH]cc1-c1ccc(cc1)-c1ccc(F)cc1 Show InChI InChI=1S/C21H14ClFN2O2/c22-18-9-16(19(26)10-20(18)27)21-17(11-24-25-21)14-3-1-12(2-4-14)13-5-7-15(23)8-6-13/h1-11,26-27H,(H,24,25) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 1.55E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research
Curated by ChEMBL
| Assay Description
Inhibition of yeast Hsp90 ATPase activity |
Bioorg Med Chem Lett 15: 3338-43 (2005)
Article DOI: 10.1016/j.bmcl.2005.05.046
BindingDB Entry DOI: 10.7270/Q22808T2 |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM50409996
(CHEMBL191228)Show InChI InChI=1S/C17H13ClN2O4/c18-12-6-10(13(21)7-14(12)22)17-11(8-19-20-17)9-1-2-15-16(5-9)24-4-3-23-15/h1-2,5-8,21-22H,3-4H2,(H,19,20) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 1.60E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research
Curated by ChEMBL
| Assay Description
Inhibition of yeast Hsp90 ATPase activity |
Bioorg Med Chem Lett 15: 3338-43 (2005)
Article DOI: 10.1016/j.bmcl.2005.05.046
BindingDB Entry DOI: 10.7270/Q22808T2 |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM50409992
(CHEMBL364882)Show InChI InChI=1S/C16H13ClN2O2/c1-9-15(10-5-3-2-4-6-10)16(19-18-9)11-7-12(17)14(21)8-13(11)20/h2-8,20-21H,1H3,(H,18,19) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 1.65E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research
Curated by ChEMBL
| Assay Description
Inhibition of yeast Hsp90 ATPase activity |
Bioorg Med Chem Lett 15: 3338-43 (2005)
Article DOI: 10.1016/j.bmcl.2005.05.046
BindingDB Entry DOI: 10.7270/Q22808T2 |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68251

(Microlactone, 16b)Show SMILES CC1C[C@H]2O[C@@H]2CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |r,w:1.0| Show InChI InChI=1S/C18H21ClO6/c1-9-6-15-14(25-15)5-3-2-4-10(20)7-11-16(18(23)24-9)12(21)8-13(22)17(11)19/h8-9,14-15,21-22H,2-7H2,1H3/t9?,14-,15-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 1.90E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68259

(Microlactone, 15e)Show SMILES C[C@@H]1CCC=CCCCC(=O)Cc2c(Cl)c(O)cc(O)c2C(=O)O1 |r,w:5.5,3.3| Show InChI InChI=1S/C18H21ClO5/c1-11-7-5-3-2-4-6-8-12(20)9-13-16(18(23)24-11)14(21)10-15(22)17(13)19/h2-3,10-11,21-22H,4-9H2,1H3/t11-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 1.90E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68258
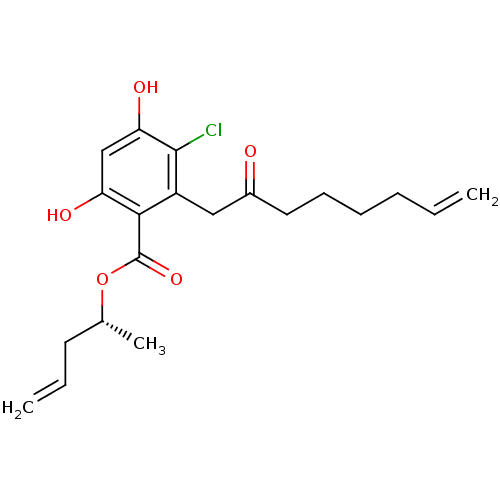
(Microlactone, 19)Show SMILES C[C@H](CC=C)OC(=O)c1c(O)cc(O)c(Cl)c1CC(=O)CCCCC=C |r| Show InChI InChI=1S/C20H25ClO5/c1-4-6-7-8-10-14(22)11-15-18(16(23)12-17(24)19(15)21)20(25)26-13(3)9-5-2/h4-5,12-13,23-24H,1-2,6-11H2,3H3/t13-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 2.10E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68249

(Microlactone, 16a)Show SMILES Oc1cc(O)c2c(CC(=O)CCCC[C@H]3O[C@@H]3CCOC2=O)c1Cl |r| Show InChI InChI=1S/C17H19ClO6/c18-16-10-7-9(19)3-1-2-4-13-14(24-13)5-6-23-17(22)15(10)11(20)8-12(16)21/h8,13-14,20-21H,1-7H2/t13-,14-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 2.10E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM20732
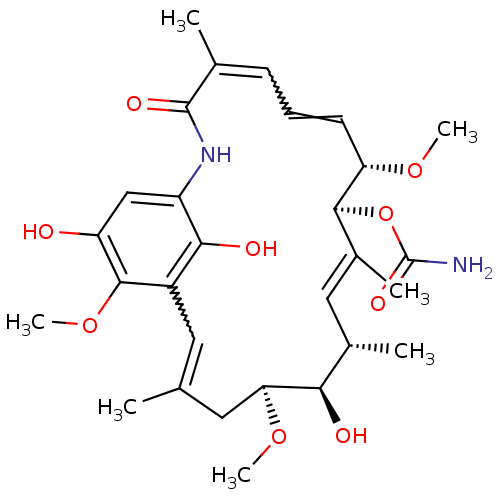
((4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14,...)Show SMILES CO[C@H]1CC(C)=Cc2c(O)c(NC(=O)C(C)=CC=C[C@H](OC)[C@@H](OC(N)=O)\C(C)=C\[C@H](C)[C@H]1O)cc(O)c2OC |r,w:16.16,6.6,t:28| Show InChI InChI=1S/C29H40N2O9/c1-15-11-19-25(34)20(14-21(32)27(19)39-7)31-28(35)16(2)9-8-10-22(37-5)26(40-29(30)36)18(4)13-17(3)24(33)23(12-15)38-6/h8-11,13-14,17,22-24,26,32-34H,12H2,1-7H3,(H2,30,36)(H,31,35)/b10-8?,15-11?,16-9?,18-13+/t17-,22-,23-,24+,26-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | 2.30E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Seoul National University
Curated by ChEMBL
| Assay Description
Inhibition of yeast Hsp90 ATPase activity by ATPase assay using malachite malachite green colorimetric method |
Bioorg Med Chem Lett 19: 4839-42 (2009)
Article DOI: 10.1016/j.bmcl.2009.06.040
BindingDB Entry DOI: 10.7270/Q2N29X0S |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM50409990
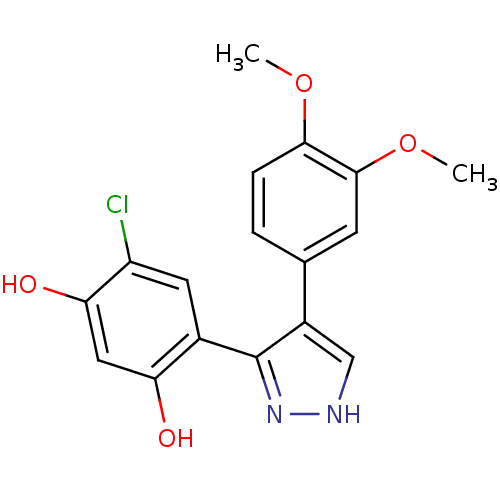
(CHEMBL190249)Show InChI InChI=1S/C17H15ClN2O4/c1-23-15-4-3-9(5-16(15)24-2)11-8-19-20-17(11)10-6-12(18)14(22)7-13(10)21/h3-8,21-22H,1-2H3,(H,19,20) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 2.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research
Curated by ChEMBL
| Assay Description
Inhibition of yeast Hsp90 ATPase activity |
Bioorg Med Chem Lett 15: 3338-43 (2005)
Article DOI: 10.1016/j.bmcl.2005.05.046
BindingDB Entry DOI: 10.7270/Q22808T2 |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM50409994

(CHEMBL188495)Show SMILES Oc1cc(O)c(cc1Cl)-c1n[nH]cc1-c1cccc(OCCCC#N)c1 Show InChI InChI=1S/C19H16ClN3O3/c20-16-9-14(17(24)10-18(16)25)19-15(11-22-23-19)12-4-3-5-13(8-12)26-7-2-1-6-21/h3-5,8-11,24-25H,1-2,7H2,(H,22,23) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 2.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research
Curated by ChEMBL
| Assay Description
Inhibition of yeast Hsp90 ATPase activity |
Bioorg Med Chem Lett 15: 3338-43 (2005)
Article DOI: 10.1016/j.bmcl.2005.05.046
BindingDB Entry DOI: 10.7270/Q22808T2 |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68260

(Microlactone, 15f cis)Show SMILES Oc1cc(O)c2c(CC(=O)CCC\C=C/CCOC2=O)c1Cl |c:13| Show InChI InChI=1S/C16H17ClO5/c17-15-11-8-10(18)6-4-2-1-3-5-7-22-16(21)14(11)12(19)9-13(15)20/h1,3,9,19-20H,2,4-8H2/b3-1- | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| DrugBank
MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 2.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68249

(Microlactone, 16a)Show SMILES Oc1cc(O)c2c(CC(=O)CCCC[C@H]3O[C@@H]3CCOC2=O)c1Cl |r| Show InChI InChI=1S/C17H19ClO6/c18-16-10-7-9(19)3-1-2-4-13-14(24-13)5-6-23-17(22)15(10)11(20)8-12(16)21/h8,13-14,20-21H,1-7H2/t13-,14-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 2.50E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68265

(Microlactone, 18)Show SMILES Oc1c(Br)c(O)c2c(CC(=O)CCCCCC=CCCOC2=O)c1Br |w:18.17,16.15| Show InChI InChI=1S/C18H20Br2O5/c19-14-12-10-11(21)8-6-4-2-1-3-5-7-9-25-18(24)13(12)16(22)15(20)17(14)23/h3,5,22-23H,1-2,4,6-10H2 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 2.63E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68258

(Microlactone, 19)Show SMILES C[C@H](CC=C)OC(=O)c1c(O)cc(O)c(Cl)c1CC(=O)CCCCC=C |r| Show InChI InChI=1S/C20H25ClO5/c1-4-6-7-8-10-14(22)11-15-18(16(23)12-17(24)19(15)21)20(25)26-13(3)9-5-2/h4-5,12-13,23-24H,1-2,6-11H2,3H3/t13-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 2.90E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68255

(Microlactone, 14c)Show SMILES C[C@@H]1C\C=C\CCCCC(=O)Cc2cc(O)cc(O)c2C(=O)O1 |r,t:3| Show InChI InChI=1S/C18H22O5/c1-12-7-5-3-2-4-6-8-14(19)9-13-10-15(20)11-16(21)17(13)18(22)23-12/h3,5,10-12,20-21H,2,4,6-9H2,1H3/b5-3+/t12-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 3.20E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
A colorimetric assay for the release of inorganic phosphate upon hydrolysis of ATP was used to determine the potency of Hsp 90 inhibitor against enzy... |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM68252

(Microlactone, 17)Show InChI InChI=1S/C18H23ClO5/c1-11-7-5-3-2-4-6-8-12(20)9-13-16(18(23)24-11)14(21)10-15(22)17(13)19/h10-11,21-22H,2-9H2,1H3 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 3.23E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Nottingham
| Assay Description
Binding competition assay with a fluorescent probe. |
Chem Biol 13: 1203-15 (2006)
Article DOI: 10.1016/j.chembiol.2006.09.015
BindingDB Entry DOI: 10.7270/Q2930RMM |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data