

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: CC1=C(CC(O)=O)c2cc(F)ccc2\C1=C/c1ccc(cc1)S(C)=O
InChI Key: InChIKey=MLKXDPUZXIRXEP-MFOYZWKCSA-N
PDB links: 9 PDB IDs match this monomer.
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lactoylglutathione lyase (Homo sapiens (Human)) | BDBM50012899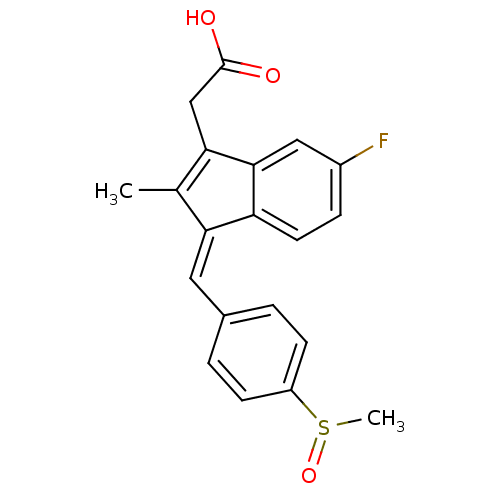 ((Z)-5-Fluoro-2-methyl-1-((p-(methylsulfinyl)phenyl...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 7.79E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sun Yat-sen University Curated by ChEMBL | Assay Description Inhibition of glyoxalase 1 | Bioorg Med Chem 19: 1189-96 (2011) Article DOI: 10.1016/j.bmc.2010.12.039 BindingDB Entry DOI: 10.7270/Q2222V2X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lactoylglutathione lyase (Homo sapiens (Human)) | BDBM50012899 ((Z)-5-Fluoro-2-methyl-1-((p-(methylsulfinyl)phenyl...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 1.39E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sun Yat-sen University Curated by ChEMBL | Assay Description Inhibition of human recombinant His-tagged glyoxalase 1 expressed in Escherichia coli BL21 (DE3) preincubated for 20 mins by Dixon plot analysis | Bioorg Med Chem 19: 1189-96 (2011) Article DOI: 10.1016/j.bmc.2010.12.039 BindingDB Entry DOI: 10.7270/Q2222V2X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Polyunsaturated fatty acid 5-lipoxygenase (Rattus norvegicus) | BDBM50012899 ((Z)-5-Fluoro-2-methyl-1-((p-(methylsulfinyl)phenyl...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | PubMed | n/a | n/a | >1.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Warner-Lambert Company Curated by ChEMBL | Assay Description Inhibition of 5-lipoxygenase in intact RBL-1 cell line | J Med Chem 33: 2070-2 (1990) BindingDB Entry DOI: 10.7270/Q2RJ4HGZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Retinoic acid receptor RXR-alpha (Homo sapiens (Human)) | BDBM50012899 ((Z)-5-Fluoro-2-methyl-1-((p-(methylsulfinyl)phenyl...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | Article PubMed | n/a | n/a | 8.29E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Xiamen University Curated by ChEMBL | Assay Description Displacement of [3H]-9-cis-RA from RXRalpha (unknown origin) by liquid scintillation counting | Eur J Med Chem 62: 632-48 (2013) Article DOI: 10.1016/j.ejmech.2013.01.012 BindingDB Entry DOI: 10.7270/Q2J104JR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Prostaglandin E synthase (Homo sapiens (Human)) | BDBM50012899 ((Z)-5-Fluoro-2-methyl-1-((p-(methylsulfinyl)phenyl...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | Article PubMed | n/a | n/a | 8.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Kentucky Curated by ChEMBL | Assay Description Inhibition of microsomal PGES | Bioorg Med Chem 19: 6077-86 (2011) Article DOI: 10.1016/j.bmc.2011.08.040 BindingDB Entry DOI: 10.7270/Q2NV9JM7 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Aldo-keto reductase family 1 member B10 [K125R,V301L] (Homo sapiens (Human)) | BDBM50012899 ((Z)-5-Fluoro-2-methyl-1-((p-(methylsulfinyl)phenyl...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | 2.69E+3 | n/a | n/a | n/a | n/a | 7.0 | n/a |
Institut de Ge´ne´tique et de Biologie Mole´culaire et Cellulaire | Assay Description The IC50-activity assays were carried out on the basis of the quantification of the NADPH consumption that takes place when the enzyme catalyzes the ... | ACS Chem Biol 11: 2693-2705 (2016) Article DOI: 10.1021/acschembio.6b00382 BindingDB Entry DOI: 10.7270/Q2NG4PFZ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Aldo-keto reductase family 1 member B1 [K65Q] (Bos taurus (Cattle)) | BDBM50012899 ((Z)-5-Fluoro-2-methyl-1-((p-(methylsulfinyl)phenyl...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | 293 | n/a | n/a | n/a | n/a | n/a | n/a |
COMSATS Institute of Information Technology | Assay Description The activity of ALR2 enzyme was determined at 340 nm in UV spectrophotometer that depends upon the measurement of NADPH consumption. Each well of the... | Bioorg Chem 68: 177-186 (2016) Article DOI: 10.1016/j.bioorg.2016.08.005 BindingDB Entry DOI: 10.7270/Q2445K96 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glycine receptor subunit alpha-1 (Homo sapiens (Human)) | BDBM50012899 ((Z)-5-Fluoro-2-methyl-1-((p-(methylsulfinyl)phenyl...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | Article PubMed | n/a | n/a | n/a | n/a | 380 | n/a | n/a | n/a | n/a |
University of Pittsburgh Curated by ChEMBL | Assay Description Potentiation of human GlyR-alpha1 expressed in Xenopus laevis oocytes assessed as induction of glycine-activated currents after 1 to 4 days by two-el... | J Med Chem 58: 2958-66 (2015) Article DOI: 10.1021/jm501873p BindingDB Entry DOI: 10.7270/Q22F7Q4V | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||